Abstract
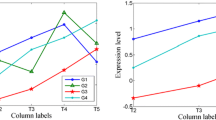
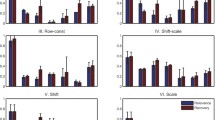
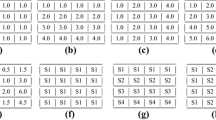
The application of high-throughput microarray has led to massive gene expression data, urging effective methodology for analysis. Biclustering comes out and serves as a useful tool, performing simultaneous clustering on rows and columns to find subsets of coherently expressed genes and conditions. Specially, in analysis of time–series gene expression data, it is meaningful to restrict biclusters to contiguous time points concerning coherent evolutions. In this paper, BCCC-Bicluster is proposed as an extension of CCC-Bicluster. An exact algorithm based on frequent sequential mining is proposed to find all maximal BCCC-Biclusters. The newly defined Frequent-Infrequent Tree-Array (FITA) is constructed to speed up the traversal process, with useful strategies originating from Apriori property to avoid redundant work. To make it more efficient, the bitwise operation XOR is applied to capture identical or opposite contiguous patterns between two rows. The algorithm is tested in simulated data, yeast microarray data and human microarray data. The experimental results show the proposed algorithm had better performance on the ability to recover the planted biclusters in the synthetic data than CCC-Biclusters and outperformed the one without FITA in speed and scalability. In the enrichment analysis, BCCC-Biclusters are proven to find more significant GO terms involved in biological processes than other three kinds of up-to-date biclusters.












Similar content being viewed by others
Explore related subjects
Discover the latest articles and news from researchers in related subjects, suggested using machine learning.References
Barkow S, Bleuler S, Prelic A, Zimmermann P, Zitzler E (2006) Bicat: a biclustering analysis toolbox. Bioinformatics 22(10):1282–1283
Ben-Dor A, Chor B, Karp R, Yakhini Z (2002) Discovering local structure in gene expression data: the order-preserving submatrix problem. In: RECOMB’02: Proceedings of the sixth annual international conference on Computational biology, pp 49–57
Bleuler S, Prelic A, Zitzler E (2004) An EA framework for biclustering of gene expression data. In: Proceedings of Congress on Evolutionary Computation, pp 166–173
Cheng Y, Church GM (2000) Biclustering of expression data. In Proceedings of the eighth international conference on intelligent systems for molecular biology, pp 93–103. AAAI Press
Cheung L, Yip KY, Cheung DW, Kao B (2007) On mining micro-array data by order-preserving submatrix. Int J Bioinform Res Appl 3:42–64
Divina F, Aguilar-Ruiz JS (2007) A multi-objective approach to discover biclusters in microarray data. In: GECCO ’07: Proceedings of the 9th annual conference on Genetic and evolutionary computation, pp 385–392
Gan XC, Liew AW, Yan H (2005) Biclustering gene expression data based on high dimensional geometric method. In: Proceedings of 4th International Conference on Machine Learning and Cybernetics, pp. 3388–3393
Gao BJ, Griffith OL, Ester M, Xiong H, Zhao Q, Jones SJM (2012) On the deep order-preserving submatrix problem: a best effort approach. IEEE Trans Knowl Data Eng 24:309–325
Getz G, Levine E, Domany E (2000) Coupled two-way clustering analysis of gene microarray data. Proc Natl Acad Sci USA 97:12079–12084
Gonçalves JP, Madeira SC, Oliveira AL (2009) BiGGEsTS: integrated environment for biclustering analysis of time series gene expression data. BMC Res Notes 2:124
Gottesman D (1998) A theory of fault-tolerant quantum computation. Phys Rev A 57, 127±137
Gu J, Liu JS (2008) Bayesian biclustering of gene expression data. BMC Genom 9(Suppl 1):S4
Hall KL, Rauschenbach KA (1998) 100-Gbit/s bitwise logic. Opt Lett 23(16):1271–1273
Hartigan JA, Wong MA (1979) A k-means Clustering Algorithm. Applied Statistics 28:100–108
Ji L, Tan KL (2005) Identifying time-lagged gene clusters using gene expression data. Bioinformatics 21:509–516
Lazzeroni L, Owen A (2002) Plaid models for gene expression data. J Statistica Sinica 12:61–86
Liu J, Yang J, Wang W (2004) Biclustering in gene expression data by tendency. In: Proceedings of Computational Systems Bioinformatics Conference, 2004. CSB 2004. IEEE. vol pp.182, 193, 16–19
Lu S, Wang X, Zhang G, Zhou X (2015) Effective algorithms of the Moore-Penrose inverse matrices for extreme learning machine. Intell Data Anal 19(4):743–760
Madeira SC, Oliveira AL (2004) Biclustering algorithms for biological data analysis: a survey. IEEE/ACM Trans Comput Biol Bioinf (TCBB) 1(1):24–45
Madeira SC, Oliveira AL (2005) A linear time biclustering algorithm for time series gene expression data. In: Proceedingsof the 5th workshop on algorithms in bioinformatics Springer Verlag, LNCS/LNBI 3692:39–52
Madeira SC, Oliveira AL (2007) An efficient biclustering algorithm for finding genes with similar patterns in time-series expression data. In: Proceedings of the 5th Asia Pacific bioinformatics conference, series in advances in bioinformatics and computational biology, vol 5. Imperial College Press, pp 67–80
Madeira SC, Teixeira MC, Sá-Correia I, Oliveira AL (2008) Identification of regulatory modules in time series gene expression data using a linear time biclustering algorithm. In: IEEE/ACM transactions on computational biology and bioinformatics, IEEE Computer Society
Madeira SC, Oliveira AL (2009) A polynomial time biclustering algorithm for finding approximate expression patterns in gene expression time series. Alg Mol Biol 4:8
Martin D, Brun C, Remy E, Mouren P, Thieffry D, Jacq B (2004) GOToolBox: functional investigation of gene datasets based on Gene Ontology. Gen Biol (12R101 [http://burgundy.cmmt.ubc.ca/GOToolBox/]
Murali TM, Kasif S (2003) Extracting conserved gene expression motifs from gene expression data. Proc Pacific Symp Biocomput 8:77–88
Peeters R (2003) The maximum edge biclique problem is NP-complete. Discrete Appl Math 131(3):651–654
Prelic A, Bleuler S, Zimmermann P, Buhlmann P, Gruissem W, Hennig L, Thiele L, Zitzler E (2006) A systematic comparison and evaluation of biclustering methods for gene expression data. Bioinformatics 22(9):1122–1129
Qu J, Zhang X, Wu L, Wang Y, Chen L (2011) Detecting coherent local patterns from time series gene expression data by a temporal biclustering method. Syst Biol (ISB), 2011 IEEE international conference on. vol pp.388, 393, 2–4
Sheng Q, Moreau Y, Moor BD (2003) Biclustering microarray data by Gibbs sampling. Bioinformatics 19(Suppl 2):196–205
Sokal RR, Michener CD (1958) A statistical method for evaluating systematic relationships. Univ Kansas Sci Bull 38:1409–1438
Tanay A, Sharan R, Shamir R (2002) Discovering statistically significant biclusters in gene expression data. Bioinformatics 18:S136–S144
Tanay A, Sharan R Shamir R (2005) Biclustering algorithms: a survey. In: Aluru S. Chapman (eds) Handbook of computational molecular biology, Hall/CRC Computer and Information Science Series
Tan KL, Eng PK, Ooi BC (2001) Efficient progressive skyline computation. In: Proceedings of the Conference on Very Large Data Bases, Rome
Wang R, Kwong S, Wang XZ, Jiang QS (2015) Segment based decision tree induction with continuous valued attributes. IEEE Trans Cybernet 45(7):1262–1275
Wang XZ, Aamir Raza Ashfaq R, Fu AM (2015) Fuzziness based sample categorization for classifier performance improvement. J Intell Fuzzy Syst 29:1185–1196
Wang XZ, Xing HJ, Li Y et al (2015) A study on relationship between generalization abilities and fuzziness of base classifiers in ensemble learning. IEEE Trans Fuzzy Syst 23(5):1638–1654
Wang XZ (2015) Uncertainty in learning from big data-editorial. J Intell Fuzzy Syst 28(5):2329–2330
Yang J, Wang H, Wang W, Yu P (2003) Enhanced biclustering on expression data. In: BIBE ’03: Proceedings of the 3rd IEEE symposium on bioinformatics and bio engineering, pp 321
Yordzhev K (2009) An example for the use of bitwise operations in programming. Math Educ Math 38:196–202
Zhang Y, Zha H, Chu CH (2005) A time-series biclustering algorithm for revealing co-regulated genes. Information technology: coding and computing, ITCC. International Conference on. vol.1, no., pp.32, 37 Vol. 1, 4–6
Zhao HY, Liew AWC, Yan H (2007) A new strategy of geometrical biclustering for microarray data analysis. In: Proc. of the Fifth Asia-Pacific Bioinformatics Conference, pp. 47–56
Acknowledgments
The authors thank gratefully for the colleagues who participated in this work and provided technical supports. This research is supported by National Natural Science Foundation of China (No. 71272084, 61572022) and the PCSIRT (Grant No. IRT1243). This work was also supported by the Scientific Research Foundation of Graduate School of South China Normal University (2015lkxm37).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interests
The authors declare that there is no conflict of interests regarding the publication of this paper.
Rights and permissions
About this article
Cite this article
Xue, Y., Ma, Z., Xu, H. et al. Discovery of bidirectional contiguous column coherent bicluster in time-series gene expression data. Int. J. Mach. Learn. & Cyber. 9, 413–426 (2018). https://doi.org/10.1007/s13042-015-0464-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13042-015-0464-0