Abstract
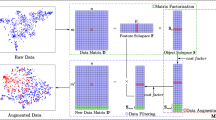
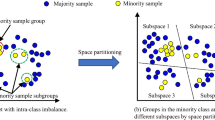
The data used for microbial-based disease diagnosis are characterized by small sample sizes, imbalanced categories, high dimensionality, and strong sparsity. They pose challenges to machine learning algorithms that aim to achieve good classification performance. In this paper, we propose a two-stage data augmentation method to enhance training data quality. The first stage is feature transformation. We design a KPCA-based method to map microbial data to a low-rank feature space, resulting in cleaner and more efficient data representation. This processing step addresses high dimensionality and strong sparsity in microbial data. The second stage is data augmentation. New synthetic data are obtained by augmenting the positive samples through the GAN. The misclassification cost is used to control the ratio of positive/negative samples in new data. The combination of the augmented data with the original data constitutes a cost-sensitive dataset, which can increase sample diversity while addressing the imbalance problem. This is more reasonable than traditional sampling methods that resolve the class imbalance. We compare the new method with four popular data augmentation algorithms on 12 imbalanced datasets. The experimental results demonstrate that (1) the samples augmented by the proposed algorithm are more diverse than those generated using compared resampling methods, such as SMOTE_ENN, and (2) the proposed algorithm not only achieves the lowest total misclassification cost but also outperforms other methods in terms of \(F_2\) and G-mean metrics.








Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.References
Ai LY, Tian HY, Chen ZF, Chen HM, Xu J, Yuan FJ (2017) Systematic evaluation of supervised classifiers for fecal microbiota-based prediction of colorectal cancer. Oncotarget 8(6):9546–9556
Asgari E, Garakani K, McHardy AC, Mofrad MR (2018) Micropheno: predicting environments and host phenotypes from 16s RRNA gene sequencing using a k-MER based representation of shallow sub-samples. Bioinformatics 34(13):i32–i42
Barandela R, Valdovinos RM, Sánchez JS (2003) New applications of ensembles of classifiers. Pattern Anal Appl 6(3):245–256
Batista GE, Bazzan AL, Monard MC et al (2003) Balancing training data for automated annotation of keywords: a case study. In: WOB, pp 10–18
Batista GE, Prati RC, Monard MC (2004) A study of the behavior of several methods for balancing machine learning training data. ACM SIGKDD Explor Newslett 6(1):20–29
Cammarota G, Ianiro G, Cianci R, Bibbò S, Gasbarrini A, Currò D (2015) The involvement of gut microbiota in inflammatory bowel disease pathogenesis: potential for therapy. Pharmacol Ther 149:191–212
Chawla NV, Bowyer KW, Hall LO, Kegelmeyer WP (2002) Smote: synthetic minority over-sampling technique. J Artif Intell Res 16:321–357
Chawla NV, Lazarevic A, Hall LO, Bowyer KW (2003) Smoteboost: improving prediction of the minority class in boosting. In: European conference on principles of data mining and knowledge discovery, Springer, pp 107–119
Chen HM, Yu Y, Wang JL, Lin YW, Kong X, Yang CQ, Yang L, Liu ZJ, Yuan YZ, Liu F, Wu JX, Zhong L, Fang DC, Zou WP, Fang JY (2013) Decreased dietary fiber intake and structural alteration of gut microbiota in patients with advanced colorectal adenoma. Am J Clin Nutr 97(5):1044–1052
Chen T, Liu X, Feng R, Wang W, Yuan C, Lu W, He H, Gao H, Ying H, Chen DZ et al (2021) Discriminative cervical lesion detection in colposcopic images with global class activation and local bin excitation. IEEE J Biomed Health Inform 26(4):1411–1421
Collado MC, Rautava S, Isolauri E, Salminen S (2015) Gut microbiota: a source of novel tools to reduce the risk of human disease? Pediatr Res 77(1):182–188
Cox LM, Blaser MJ (2015) Antibiotics in early life and obesity. Nat Rev Endocrinol 11(3):182–190
Dhar S, Cherkassky V (2014) Development and evaluation of cost-sensitive universum-SVM. IEEE Trans Cybern 45(4):806–818
Esteva A, Kuprel B, Novoa RA, Ko J, Swetter SM, Blau HM, Thrun S (2017) Dermatologist-level classification of skin cancer with deep neural networks. Nature 542(7639):115–118
Gao H, Xu K, Cao M, Xiao J, Xu Q, Yin Y (2021) The deep features and attention mechanism-based method to dish healthcare under social IOT systems: an empirical study with a hand-deep local-global net. IEEE Trans Comput Soc Syst 9(1):336–347
Gao H, Xiao J, Yin Y, Liu T, Shi J (2022) A mutually supervised graph attention network for few-shot segmentation: the perspective of fully utilizing limited samples. IEEE Trans Neural Netw Learn Syst
Gohir W, Ratcliffe EM, Sloboda DM (2015) Of the bugs that shape us: maternal obesity, the gut microbiome, and long-term disease risk. Pediatr Res 77(1):196–204
Guo SY, Rong Z, Wang S, Wu YH (2022) A lidar slam with PCA-based feature extraction and two-stage matching. IEEE Trans Instrum Meas 71:1–11
Halfvarson J, Brislawn CJ, Lamendella R, Vázquez-Baeza Y, Walters WA, Bramer LM, D’Amato M, Bonfiglio F, McDonald D, Gonzalez A, McClure EE, Dunklebarger M, Knight R, Jansson JK (2017) Dynamics of the human gut microbiome in inflammatory bowel disease. Nat Microbiolo 2(5):17004–17004
Han H, Wang WY, Mao BH (2005) Borderline-smote: a new over-sampling method in imbalanced data sets learning. In: International conference on intelligent computing, Springer, pp 878–887
He H, Bai Y, Garcia EA, Li S (2008) Adasyn: adaptive synthetic sampling approach for imbalanced learning. In: 2008 IEEE international joint conference on neural networks (IEEE world congress on computational intelligence), IEEE, pp 1322–1328
Kostic AD, Xavier RJ, Gevers D (2014) The microbiome in inflammatory bowel disease: current status and the future ahead. Gastroenterology 146(6):1489–1499
Larsen PE, Dai Y (2015) Metabolome of human gut microbiome is predictive of host dysbiosis. GigaScience 4(1):s13742-015
Last F, Douzas G, Bacao F (2017) Oversampling for imbalanced learning based on k-means and smote. arXiv preprint arXiv:1711.00837
Li Y, Huang C, Ding L, Li Z, Pan Y, Gao X (2019) Deep learning in bioinformatics: introduction, application, and perspective in the big data era. Methods 166:4–21
Li YX, Chai Y, Yin HP, Chen B (2021) A novel feature learning framework for high-dimensional data classification. Int J Mach Learn Cybern 12(2):555–569
Liu Y, Kohlberger T, Norouzi M, Dahl GE, Smith JL, Mohtashamian A, Olson N, Peng LH, Hipp JD, Stumpe MC (2019) Artificial intelligence-based breast cancer nodal metastasis detection: insights into the black box for pathologists. Arch Pathol Lab Med 143(7):859–868
Lo C, Marculescu R (2019) Metann: accurate classification of host phenotypes from metagenomic data using neural networks. BMC Bioinform 20(Suppl 12):1–14
Luo S, Chen Z (2014) Sequential lasso cum EBIC for feature selection with ultra-high dimensional feature space. J Am Stat Assoc 109(507):1229–1240
Mahindru A, Sangal A (2021) Semidroid: a behavioral malware detector based on unsupervised machine learning techniques using feature selection approaches. Int J Mach Learn Cybern 12(5):1369–1411
Mani I, Zhang I (2003) KNN approach to unbalanced data distributions: a case study involving information extraction. In: Proceedings of workshop on learning from imbalanced datasets, ICML 126, pp 1–7
Mountassir A, Benbrahim H, Berrada I (2012) An empirical study to address the problem of unbalanced data sets in sentiment classification. In: 2012 IEEE international conference on systems, man, and cybernetics (SMC), IEEE, pp 3298–3303
Pasolli E, Truong DT, Malik F, Waldron L, Segata N (2016) Machine learning meta-analysis of large metagenomic datasets: tools and biological insights. PLoS Comput Biol 12(7):e1004977
Reiman D, Metwally AA, Dai Y (2017) Using convolutional neural networks to explore the microbiome. In: 2017 39th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), pp 4269–4272
Reshef DN, Reshef YA, Finucane HK, Grossman SR, McVean G, Turnbaugh PJ, Lander ES, Mitzenmacher M, Sabeti PC (2011) Detecting novel associations in large data sets. Science 334(6062):1518–1524
Rosipal R, Girolami M, Trejo LJ, Cichocki A (2001) Kernel PCA for feature extraction and de-noising in nonlinear regression. Neural Comput Appl 10(3):231–243
Sahin Y, Bulkan S, Duman E (2013) A cost-sensitive decision tree approach for fraud detection. Expert Syst Appl 40(15):5916–5923
Seiffert C, Khoshgoftaar TM, Van Hulse J, Napolitano A (2009) Rusboost: a hybrid approach to alleviating class imbalance. IEEE Trans Syst Man Cybern Part A: Syst Hum 40(1):185–197
Van’t Veer LJ, Dai H, Van De Vijver MJ, He YD, Hart AA, Mao M, Peterse HL, Van Der Kooy K, Marton MJ, Witteveen AT et al (2002) Gene expression profiling predicts clinical outcome of breast cancer. Nature 415(6871):530–536
Wang S, Yao X (2009) Diversity analysis on imbalanced data sets by using ensemble models. In: 2009 IEEE symposium on computational intelligence and data mining, IEEE, pp 324–331
Wen LY, Luo CG, Wu WZ, Min F (2020) Multi-label symbolic value partitioning through random walks. Neurocomputing 387:195–209
Wilson DL (1972) Asymptotic properties of nearest neighbor rules using edited data. IEEE Trans Syst Man Cybern 3:408–421
Wu HL, Cai LH, Li DF, Wang XY, Zhao SC, Zou FH, Zhou K (2018) Metagenomics biomarkers selected for prediction of three different diseases in Chinese population. Biomed Res Int 2018:1–7
Wu J, Wang J, Liu L (2007) Feature extraction via KPCA for classification of gait patterns. Hum Mov Sci 26(3):393–411
Yang LY, Xu ZS (2019) Feature extraction by PCA and diagnosis of breast tumors using SVM with de-based parameter tuning. Int J Mach Learn Cybern 10(3):591–601
Ye MC, Ji CX, Chen H, Lei L, Lu HJ, Qian YT (2020) Residual deep PCA-based feature extraction for hyperspectral image classification. Neural Comput Appl 32(18):14287–14300
Zhang X, Yang Y, Li T, Zhang Y, Wang H, Fujita H (2021) CMC: a consensus multi-view clustering model for predicting Alzheimer’s disease progression. Comput Methods Programs Biomed 199:105895
Zhang Y, Zhang HP (2013) Microbiota associated with type 2 diabetes and its related complications. Food Sci Hum Wellness 2(3–4):167–172
Zhang ZL, Luo XG, García S, Herrera F (2017) Cost-sensitive back-propagation neural networks with binarization techniques in addressing multi-class problems and non-competent classifiers. Appl Soft Comput 56:357–367
Acknowledgements
This work was supported by the Central Government Funds of Guiding Local Scientific and Technological Development (No. 2021ZYD0003) and the Scientific Research Starting Project of SWPU (No. 2018QHR007).
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wen, LY., Zhang, XM., Li, QF. et al. KGA: integrating KPCA and GAN for microbial data augmentation. Int. J. Mach. Learn. & Cyber. 14, 1427–1444 (2023). https://doi.org/10.1007/s13042-022-01707-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13042-022-01707-3