Abstract
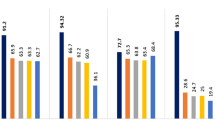
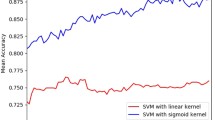
Proteins have crucial importance in every living system, and Protein–Protein Interactions (PPIs) play a pivotal role in regulation of virtually all biological processes such as DNA transcription, replication, metabolic cycles and signaling cascades. The PPIs play an important role in the complex process of cell death which mainly occurs via apoptosis and necrosis in eukaryotic cells. Apoptosis is an orderly cellular suicide program critical for the development and homeostasis of multicellular organism. Failure to control apoptosis can have catastrophic consequences. The cascades of amazing reactions carried out by proteins such as Caspase, CARD, NLRP, NOD, FADD, DEDD, POP, Myd88 etc. play important role in the process of cell death. The high throughput experimental methods for determining PPIs are time consuming, expensive and are generating huge amount of PPIs data. Therefore, there is need to develop computational methods to efficiently and accurately predict PPIs. In this work, an attempt has been made to develop a fuzzy support vector machine (F-SVM) model for predicting death domain (DD) PPIs based on sixteen physicochemical, biochemical and structural features of amino acids which are monomers of proteins. First, the protein primary sequences are encoded into sequential features represented by descriptors. Then, the Support Vector Machine and Sequential Minimal Optimization of WEKA software are employed to classify interacting and non-interacting protein pairs. The performance of SVM and F-SVM with various kernel functions has been evaluated and it was observed that libSVM with Polynomial kernel was found to be best with accuracy of 77.94 % via F-SVM which is optimum model in predicting human DD-PPIs. Validation is performed by tenfold cross-validation technique. The F-SVM performance measure is 2.94 % higher than SVM in terms of accuracy with the use of custom designed fuzzy membership function. The results obtained are in agreement with available experimental data. Such models can be useful in providing PPI information of DD proteins which can be useful in understanding the molecular mechanisms involved in death of cells taking place due to aging, programmed cell death and various diseases. It may through some light on the study of cancerous cell and gerontology.




Similar content being viewed by others
Abbreviations
- PPIs:
-
Protein—protein interactions
- F-SVM:
-
Fuzzy—support vector machine
- SMO:
-
Sequential minimal optimization
- MCC:
-
Mathew correlation coefficient
- Apaf-1:
-
Apoptotic protease activating factor-1
- ASC:
-
Apoptosis associated Spec-like protein containing a CARD
- Bcl-10:
-
B cell lymphoma/leukemia 10
- BinCARD:
-
Bcl-10 interacting CARD protein
- CARD:
-
Caspase-recruitment domain
- Caspase:
-
Cysteine aspartate-specific proteinase
- CIAP:
-
Baculoviral IAP repeat containing protein
- ANK3:
-
Ankyrin 3
- DD:
-
Death domain
- DAPK:
-
Death-associated protein kinase
- DED:
-
Death effector domain
- DR:
-
Tumor necrosis factor receptor superfamily member
- FADD:
-
Fas-associated death domain
- IRAK:
-
Interleukin receptor associated kinase
- MAVS:
-
Mitochondrial antiviral signaling protein
- Myd88:
-
Myeloid differentiation primary response protein MyD88
- NLRP:
-
NACHT LRR and PYD domain containing protein
- NOD:
-
Nucleotide binding oligomerization domain protein
- POP:
-
Pyrin domain containing protein
- RAIDD:
-
Death domain containing protein CRADD
- RIG1:
-
Probable ATP dependent RNA helicase
- RIPK:
-
Receptor interacting Ser/Thr protein kinase
References
Bock JR, Gough DA (2001) Predicting protein–protein interactions from primary structure. Bioinformatics 17:455–460
Bratton SB, Walker G, Srinivasula SM, Sun XM, Butterworth M, Alnemri ES, Cohen GM (2001) Recruitment, activation and retention of caspases-9 and-3 by Apaf-1 apoptosome and associated XIAP complexes. EMBO J 20:998–1009
Candi E, Schmidt R, Melino G (2005) The cornified envelope: a model of cell death in the skin. Nat Rev Mol Cell Biol 6:328–340
Chang CC, Lin C-J (2011) LIBSVM: a library for support vector machines. ACM Trans Intell Syst Technol (TIST) 2:27
Chothia C, Janin J (1975) Principles of protein-protein recognition. Nature 256:705–708
Coussens LM, Werb Z (2002) Inflammation and cancer Nature 420:860–867
Cristianini N, Shawe-Taylor J (2000) An introduction to support vector machines and other kernel-based learning methods. Cambridge University Press, Cambridge
Curtin JF, Cotter TG (2002) Apoptosis: historical perspectives. Essays Biochem 39:1–10
Eckhart L, Lippens S, Tschachler E, Declercq W (2013) Cell death by cornification. Biochim Biophys Acta (BBA)-Mol Cell Res 1833:3471–3480
Ferri C, Hernández-Orallo J, Modroiu R (2009) An experimental comparison of performance measures for classification Pattern Recognition Letters 30:27–38
Fields S, Song O-k (1989) A novel genetic system to detect protein–protein interactions. Nature 340:245–246
Gavin A-C et al (2002) Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415:141–147
Gavin A-C et al (2006) Proteome survey reveals modularity of the yeast cell machinery. Nature 440:631–636
Golstein P, Kroemer G (2007) Cell death by necrosis: towards a molecular definition. Trends Biochem Sci 32:37–43
Guo Y, Yu L, Wen Z, Li M (2008) Using support vector machine combined with auto covariance to predict protein–protein interactions from protein sequences. Nucleic Acids Res 36:3025–3030
Hall M, Frank E, Holmes G, Pfahringer B, Reutemann P, Witten IH (2009) The WEKA data mining software: an update. ACM SIGKDD Explor Newsl 11:10–18
Hanley JA, McNeil BJ (1982) The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology 143:29–36
Ho Y et al (2002) Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415:180–183
Holmes G, Donkin A, Witten IH Weka: A machine learning workbench. In: Intelligent Information Systems, 1994. In: Proceedings of the 1994 Second Australian and New Zealand Conference on, 1994. IEEE, pp 357–361
Huang J, Ling CX (2005) Using AUC and accuracy in evaluating learning algorithms. Knowl Data Eng IEEE Trans 17:299–310
Ito T, Chiba T, Ozawa R, Yoshida M, Hattori M, Sakaki Y (2001) A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc Natl Acad Sci 98:4569–4574
Jones S, Thornton JM (1996) Principles of protein-protein interactions. Proc Natl Acad Sci 93:13–20
Kawashima S, Ogata H, Kanehisa M (1999) AAindex: amino acid index database. Nucleic Acids Res 27:368–369
Kerr JF, Wyllie AH, Currie AR (1972) Apoptosis: a basic biological phenomenon with wide-ranging implications in tissue kinetics. Br J Cancer 26:239
Kroemer G, Galluzzi L, Vicencio JM, Kepp O, Tasdemir E, Maiuri MC (2008) To die or not to die: that is the autophagic question. Curr Mol Med 8:78–91
Kwon D et al (2012) A comprehensive manually curated protein–protein interaction database for the Death Domain superfamily. Nucleic Acids Res 40:D331–D336
Levine B, Yuan J (2005) Autophagy in cell death: an innocent convict? J Clin Investig 115:2679–2688
Lin C-F, Wang S-D (2002) Fuzzy support vector machines. Neural Netw IEEE Trans 13:464–471
Marcotte EM, Pellegrini M, Ng H-L, Rice DW, Yeates TO, Eisenberg D (1999) Detecting protein function and protein-protein interactions from genome sequences. Science 285:751–753
Martin SJ, Green DR (1995) Protease activation during apoptosis: death by a thousand cuts? Cell 82:349–352
Martin S, Roe D, Faulon J-L (2005) Predicting protein–protein interactions using signature products. Bioinformatics 21:218–226
Mehrpour M, Esclatine A, Beau I, Codogno P (2010) Autophagy in health and disease. 1. Regulation and significance of autophagy: an overview. Am J Physiol Cell Physiol 298:C776–C785
Melino G (2001) The Sirens’ song. Nature 412:23
Oncley J, Ellenbogen E, Gitlin D, Gurd F (1952) Protein–protein interactions. J Phys Chem 56:85–92
Phizicky EM, Fields S (1995) Protein-protein interactions: methods for detection and analysis. Microbiol Rev 59:94–123
Platt JC (1999) Fast training of support vector machines using sequential minimal optimization. Advances in kernel methods. MIT press, Cambridge, pp 185–208
Proskuryakov SY, Konoplyannikov AG, Gabai VL (2003) Necrosis: a specific form of programmed cell death? Exp Cell Res 283:1–16
Reed JC, Doctor KS, Godzik A (2004) The domains of apoptosis: a genomics perspective. Sci Signal 2004:re9
Rieux-Laucat F, Fischer A, Deist FL (2003) Cell-death signaling and human disease. Curr Opin Immunol 15:325–331
Shah AR, Oehmen CS, Webb-Robertson B-J (2008) SVM-HUSTLE—an iterative semi-supervised machine learning approach for pairwise protein remote homology detection. Bioinformatics 24:783–790
Shen J et al (2007) Predicting protein–protein interactions based only on sequences information. Proc Natl Acad Sci 104:4337–4341
Valencia A, Pazos F (2002) Computational methods for the prediction of protein interactions. Curr Opin Struct Biol 12:368–373
Vapnik V (1998) Statistical learning theory. 1998. Wiley, New York
Von Mering C, Krause R, Snel B, Cornell M, Oliver SG, Fields S, Bork P (2002) Comparative assessment of large-scale data sets of protein–protein interactions. Nature 417:399–403
Waugh DF (1954) Protein-protein interactions. Adv Protein Chem 9:325–437
White E (1993) Death-defying acts: a meeting review on apoptosis. Genes Dev 7:2277–2284
Witten IH, Frank E (2005) Data Mining: Practical machine learning tools and techniques. Morgan Kaufmann
Yang L, Xia J-F, Gui J (2010) Prediction of protein-protein interactions from protein sequence using local descriptors. Protein Pept Lett 17:1085–1090
Zaki N, Deris S, Alashwal H (2006) Protein-protein interaction detection based on substring sensitivity measure. Inter J Biomed Sci 1:148–154
Acknowledgments
Authors are thankful to Bioinformatics Infrastructure Facility (BIF) of Department of Biotechnology (DBT), Ministry of Science and Technology, Government of India.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Nemade, P.A., Pardasani, K.R. Fuzzy support vector machine model to predict human death domain protein–protein interactions. Netw Model Anal Health Inform Bioinforma 4, 5 (2015). https://doi.org/10.1007/s13721-015-0078-1
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13721-015-0078-1