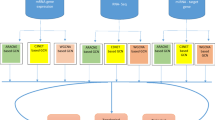
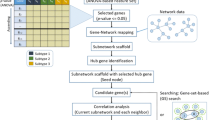
Abstract
Selection of important genes responsible for a disease is an important task in bioinformatics. Microarray data are often used with differential expression being considered as a cue. Recently, such expression data are supplemented by gene ontology and genes/proteins interaction network for the selection task. The functional knowledge and interaction structure have become critical for understanding the biological processes, including selection of genes potentially associated to complex diseases. In this paper, we propose an approach that combines expression analysis with structural analysis of protein–protein interaction networks to identify genes associated with complex diseases. The dense subgraph structures embedded in the networks are extracted. We present results on three different types of benchmark cancer dataset (prostate cancer, interstitial lung disease and chronic lymphocytic leukemia) and show that several interesting biological information could be inferred, besides achieving a high prediction accuracy. The proposed methodology helps to identify not just differentially expressed genes but also hub genes important in biological processes.











Similar content being viewed by others
References
Ahn J, Yoon Y, Park C, Shin E, Park S (2011) Integrative gene network construction for predicting a set of complementary prostate cancer genes. Bioinformatics 27(13):1846–1853
Archer SY, Hodint RA (1999) Histone acetylation and cancer. Curr Opin Genet Dev 9(2):171–174
Barabási AL, Gulbahce N, Loscalzo J (2011) Network medicine: a network-based approach to human disease. Nat Rev Genet 12(1):56–68
Berriz GF, Beaver JE, Cenik C, Tasan M, Roth FP (2009) Next generation software for functional trend analysis. Bioinformatics 25(22):3043–3044
Blondel VD, Guillaume JL, Lambiotte R, Lefebvre E (2008) Fast unfolding of communities in large networks. J Stat MechTheor Exp 2008(10):P10,008
Braoudaki M, Lambrou GI, Vougas K, Karamolegou K, Tsangaris GT, Tzortzatou-Stathopoulou F (2013) Protein biomarkers distinguish between high-and low-risk pediatric acute lymphoblastic leukemia in a tissue specific manner. J Hematol Oncol 6(1):52
Calvano SE, Xiao W, Richards DR, Felciano RM, Baker HV, Cho RJ, Chen RO, Brownstein BH, Cobb JP, Tschoeke SK et al (2005) A network-based analysis of systemic inflammation in humans. Nature 437(7061):1032–1037
Cho JH, Gelinas R, Wang K, Etheridge A, Piper MG, Batte K, Dakhlallah D, Price J, Bornman D, Zhang S et al (2011) Systems biology of interstitial lung diseases: integration of mrna and microrna expression changes. BMC Med Genom 4(1):8
Chuang HY, Lee E, Liu YT, Lee D, Ideker T (2007) Network-based classification of breast cancer metastasis. Mol Syst Biol 3(1)
Consortium TGO(2015) Gene ontology consortium: going forward. Nucl Acids Res 43D1:D1049–D1056
Cun Y, Fröhlich H (2012) Biomarker gene signature discovery integrating network knowledge. Biology 1(1):5–17
Cun Y, Fröhlich H (2013) Network and data integration for biomarker signature discovery via network smoothed t-statistics. PloS one 8(9):e73,074
Dao P, Wang K, Collins C, Ester M, Lapuk A, Sahinalp SC (2011) Optimally discriminative subnetwork markers predict response to chemotherapy. Bioinformatics 27(13):i205–i213
Díaz-Uriarte R, De Andres SA (2006) Gene selection and classification of microarray data using random forest. BMC Bioinf 7(1):3
Dittrich MT, Klau GW, Rosenwald A, Dandekar T, Müller T (2008) Identifying functional modules in protein-protein interaction networks: an integrated exact approach. Bioinformatics 24(13):i223–i231
Enright AJ, Iliopoulos I, Kyrpides NC, Ouzounis CA (1999) Protein interaction maps for complete genomes based on gene fusion events. Nature 402(6757):86–90
Fält S, Merup M, Gahrton G, Lambert B, Wennborg A (2005) Identification of progression markers in b-cll by gene expression profiling. Experim Hematol 33(8):883–893
Feng J, Jiang R, Jiang T (2011) A max-flow-based approach to the identification of protein complexes using protein interaction and microarray data. Comput Biol Bioinf IEEE/ACM Trans 8(3):621–634
Fortney K, Kotlyar M, Jurisica I (2010) Method inferring the functions of longevity genes with modular subnetwork biomarkers of caenorhabditis elegans aging. Genom Biol R13
Fortunato S (2010) Community detection in graphs. Phys Rep 486(3):75–174
Gaiteri C, Ding Y, French B, Tseng GC, Sibille E (2014) Beyond modules and hubs: the potential of gene coexpression networks for investigating molecular mechanisms of complex brain disorders. Genes Brain Behav 13(1):13–24
Ghosh A, Dhara BC, De RK (2014) Selection of genes mediating certain cancers, using a neuro-fuzzy approach. Neurocomputing 133:122–140
Goh KI, Cusick ME, Valle D, Childs B, Vidal M, Barabási AL (2007) The human disease network. Proc Natl Acad Sci 104(21):8685–8690
Goldberg AV (1984) Finding a maximum density subgraph. University of California Berkeley, CA
Guo Z, Zhang T, Li X, Wang Q, Xu J, Yu H, Zhu J, Wang H, Wang C, Topol EJ et al (2005) Towards precise classification of cancers based on robust gene functional expression profiles. BMC Bioinform 6(1):58
Hanahan D, Weinberg RA (2011) Hallmarks of cancer: the next generation. Cell 144(5):646–674
He L, Wang Y, Yang Y, Huang L, Wen Z (2014) Identifying the gene signatures from gene-pathway bipartite network guarantees the robust model performance on predicting the cancer prognosis. BioMed Res Int 2014:424509
Hu H, Yan X, Huang Y, Han J, Zhou XJ (2005) Mining coherent dense subgraphs across massive biological networks for functional discovery. Bioinformatics 21(suppl 1):i213–i221
Huang DW, Sherman BT, Lempicki RA (2008) Systematic and integrative analysis of large gene lists using david bioinformatics resources. Nat Protoc 4(1):44–57
Huang DW, Sherman BT, Lempicki RA (2009) Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucl Acids Res 37(1):1–13
Ideker T, Sharan R (2008) Protein networks in disease. Genome Res 18(4):644–652
Ji J, Zhang A, Liu C, Quan X, Liu Z (2014) Survey: Functional module detection from protein-protein interaction networks. Knowl Data Eng IEEE Trans 26(2):261–277
Kerrien S, Aranda B, Breuza L, Bridge A, Broackes-Carter F, Chen C, Duesbury M, Dumousseau M, Feuermann M, Hinz U, Jandrasits C, Jimenez RC, Khadake J, Mahadevan U, Masson P, Pedruzzi I, Pfeiffenberger E, Porras P, Raghunath A, Roechert B, Orchard S, Hermjakob H (2012) The intact molecular interaction database in 2012. Nucl Acids Res 40(Database issue):D841–D846
Langfelder P, Horvath S (2008) Wgcna: an r package for weighted correlation network analysis. BMC Bioinform 9(1):559
Lee VE, Ruan N, Jin R, Aggarwal C (2010) A survey of algorithms for dense subgraph discovery. In: Managing and Mining Graph Data, vol 40. Springer, Heidelberg, pp 303–336
Licata L, Briganti L, Peluso D, Perfetto L, Iannuccelli M, Galeota E, Sacco F, Palma A, Nardozza AP, Santonico E, Castagnoli L, Cesareni G (2012) MINT, the molecular interaction database: 2012 update. Nucl Acids Res 40(Database issue):D857–D861. doi:10.1093/nar/gkr930
Ma X, Gao L (2012) Predicting protein complexes in protein interaction networks using a core-attachment algorithm based on graph communicability. Inf Sci 189:233–254
Ma Y, Gal A, Koss MN (2007) The pathology of pulmonary sarcoidosis: update. In: Seminars in diagnostic pathology, vol 24. Elsevier, pp. 150–161
Masdehors P, Merle-Béral H, Magdelénat H, Delic J (2000) Ubiquitin-proteasome system and increased sensitivity of b-cll lymphocytes to apoptotic death activation. Leuk Lymph 38(5–6):499–504
Mertens D, Stilgenbauer S (2014) Prognostic and predictive factors in patients with chronic lymphocytic leukemia: Relevant in the era of novel treatment approaches? J Clin Oncol 32(9):869–872
Mesri EA, Feitelson MA, Munger K (2014) Human viral oncogenesis: A cancer hallmarks analysis. Cell Host Microb 15(3):266–282
Mitra K, Carvunis AR, Ramesh SK, Ideker T (2013) Integrative approaches for finding modular structure in biological networks. Nat Rev Genet 14(10):719–732
Park C, Ahn J, Kim H, Park S (2014) Integrative gene network construction to analyze cancer recurrence using semi-supervised learning. PloS One 9(1):e86,309
Prasad TSK, Goel R, Kandasamy K, Keerthikumar S, Kumar S, Mathivanan S, Telikicherla D, Raju R, Shafreen B, Venugopal A (2009) Human Protein Reference Database—2009 update. Nucl Acids Res 37(Database issue):D767–D772
Rahmani H, Blockeel H, Bender A (2011) Interaction-based feature selection for predicting cancer-related proteins in protein-protein interaction networks. In: Prooceedings of the Fifth International Workshop on Machine Learning in Systems Biology, pp 68–72
Rapaport F, Zinovyev A, Dutreix M, Barillot E, Vert JP (2007) Classification of microarray data using gene networks. BMC Bioinform 8(1):35
Ren X, Wang Y, Zhang XS, Jin Q (2013) ipcc: a novel feature extraction method for accurate disease class discovery and prediction. Nucl Acids Res gkt343
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13(11):2498–2504
Soh D, Dong D, Guo Y, Wong L (2011) Finding consistent disease subnetworks across microarray datasets. BMC Bioinform 12(Suppl 13):S15
Sterner DE, Berger SL (2000) Acetylation of histones and transcription-related factors. Microbiol Mol Biol Rev 64(2):435–459
Stuart JM, Segal E, Koller D, Kim SK (2003) A gene-coexpression network for global discovery of conserved genetic modules. Science (New York, N.Y.) 302(5643):249–255. doi:10.1126/science.1087447
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES et al (2005) Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 102(43):15545–15550
Swarnkar T, Simões SN, Martins-Jr, DC, Anura A, Brentani H, Hashimoto RF, Mitra P (2014) Multiview clustering on ppi network for gene selection and enrichment from microarray data. In: IEEE international conference on bioinformatics and bioengineering, pp 15–22
Taylor IW, Linding R, Warde-Farley D, Liu Y, Pesquita C, Faria D, Bull S, Pawson T, Morris Q, Wrana JL (2009) Dynamic modularity in protein interaction networks predicts breast cancer outcome. Nat Biotechnol 27(2):199–204
Tsourakakis C, Bonchi F, Gionis A, Gullo F, Tsiarli M (2013) Denser than the densest subgraph: extracting optimal quasi-cliques with quality guarantees. In: Proceedings of the 19th ACM SIGKDD international conference on Knowledge discovery and data mining. Chicago, pp 104–112
Wu C, Zhu J, Zhang X (2012) Integrating gene expression and protein-protein interaction network to prioritize cancer-associated genes. BMC Bioinform 13(1):182
Wu X, Jiang R, Zhang MQ, Li S (2008) Network-based global inference of human disease genes. Mol Syst Biol 4(1):189
Xiao Y, Hsiao TH, Suresh U, Chen HIH, Wu X, Wolf SE, Chen Y (2014) A novel significance score for gene selection and ranking. Bioinformatics 30(6):801–807
Xu X, Zhang A (2005) Selecting informative genes from microarray dataset by incorporating gene ontology. In: Fifth IEEE Symposium on Bioinformatics and Bioengineering, 2005. BIBE 2005. pp 241–245
Zhang W, Sun F, Jiang R (2011) Integrating multiple protein–protein interaction networks to prioritize disease genes: a bayesian regression approach. BMC Bioinform 12(Suppl 1):S11
Zhu Y, Shen X, Pan W (2009) Network-based support vector machine for classification of microarray samples. BMC Bioinform 10(Suppl 1):S21
Acknowledgments
We would like to thank the financial support from CAPES, CNPq, FAPESP (Grant 2011/50761-2), FAPESP-Microsoft (Grant 2010/52138-8), eScience-PRP-USP and Indo-Brazil Collaborative Project, DST, Govt. of India and Govt. of Brazil.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest and that this research did not involve human participants and/or animals.
Rights and permissions
About this article
Cite this article
Swarnkar, T., Simões, S.N., Anura, A. et al. Identifying dense subgraphs in protein–protein interaction network for gene selection from microarray data. Netw Model Anal Health Inform Bioinforma 4, 33 (2015). https://doi.org/10.1007/s13721-015-0104-3
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13721-015-0104-3