Abstract
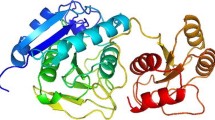
Microbial glutaminase has been extensively used as antitumor drug in pharmaceutical industry from past few decades. The structural analysis based on homology modeling predicted a hypothetical 3D model structure for l-glutaminase from Bacillus cereus MTCC 1305 using peptide sequence data obtained from MALDI-TOF MS and template model of X-ray crystal structure of l-glutaminase from Bacillus subtilis (PDB: 1MKI_A). The model was found more reliable with 98.8 % residues in the favored region of Ramachandran plot. The predicted model structure was further refined using ANOLEA, QMEAN score and GROMOS 96 force field with total energy of −11,636.01 kJ/Mol and Z-score value as −1.41. Active site of predicted model was found to be enriched with 11 conserved amino acid residues like Ser73, ASN125, Asn176, Val270, Gln72, Cys204, Val270, Ser271, Tyr200, Lys76, Phe105, Tyr252. The docking approach showed good binding affinity of predicted model towards l-glutamine with favorable ΔG docking score. The anticancer activity of this enzyme was confirmed against colon carcinoma (HCT-116) cell line with IC50 value of 99.79 μg/ml.



Similar content being viewed by others
References
Aladdin A, Bouna A, Ibrahim A, Noor RM, Ramli F, Shamsir MS (2014) Homology modeling and molecular dynamics simulation of a novel β-galactosidase from antarctic psychrophilic bacterium planococcus antarcticus DSM 14505. J Biotechnol Sci Res 2(1):63–70
Arnold K, Bordoli L, Kopp J, Schwede T (2006) The SWISS-MODEL workspace: a web-based environment for protein structure homology modeling. Bioinformatics 22:195–201
Bachmair A, Finley D, Varshavsky A (1986) In vivo half-life of a protein is a function of its amino-terminal residue. Science 234:179–186
Baig MS, Manickem N (2010) Homology modeling and docking studies of Comamonas testostroni B-356 biphenyl-2,3-dioxygenase involved in degradation of polychlorinated biphenyls. Int J Biol Macromol 46:47–53
Beedkar SD, Khobragade CN, Bodade RG, Vinchurkar AS (2012) Comparative structural modeling and docking studies of uricase: possible implication in enzyme supplementation therapy for hyperuricemic disorders. Comput Biol Med 42:657–666
Bodade RG, Beedkar SD, Manwar AV, Khobragade CN (2010) Homology modeling and docking study of xanthine oxidase of Arthobacter sp. XL 26. Int J Biol Macromol 47:298–303
Brown G, Singer A, Dementieva I, Proudfoot M, Kuznetsova E, Skarina T, Gonzalez CF, Kim YC, Joachimiak A, Chang C, Savchenko A, Yakunin AF (2008) Functional and structural characterization of four glutaminases from Escherichia coli and Bacillus subtilis. Biochem 47:5724–5735
Cappelletti D, Chiarelli LR, Pasquetto MV, Stivala CV, Scotti C (2008) Helicobacter pylori Lasparaginase: a promising chemotherapeutic agent. Biochem Biophys Res Commun 377:1222–1226
Edelhoch H (1967) Spectroscopic determination of tryptophan and tyrosine in proteins. Biochem 6:1948–1954
El-Ghonemy DH (2014) Microbial amidases and their industrial applications: a review. J Med Microb Diagn 4:173. doi:10.4172/21610703.1000173
Elshafei AM, Hassan MM, Abouzeid MA, Mahmoud DA, El-Ghonemy DH (2014) Purification, kinetic properties and antitumor activity of l-glutaminase from Penicillium brevicompactum NRC 829. Br Microbiol Res J 4:93–111
Gallagher MP, Marshall RD, Wilson R (1989) Asparaginase as a drug for treatment of acute lyphoblastic leukaemia. Essays Biochem 24:1–40
Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, Bairoch A (2005). Protein identification and analysis tools on the ExPASy server. In: Walker JM (eds) The proteomics protocols handbook. Totowa, NJ, pp 571–607
Gill SC, Von Hippel PH (1989) Calculation of protein extinction coefficients from amino acid sequence data. Anal Biochem 182:319–326
Gonda DK, Bachmair A, Wunning I, Tobias JW, Lane WS, Varshavsky AJ (1989) Universality and structure of the N-end rule. J Biol Chem 264:16700–16712
Grosdidier S, Totrov M, Fernandez RJ (2009) Computer applications for prediction of protein-protein interactions and rational drug design. Adv App Bioinf Chem 2:101–123
Guex N, Peitsch MC (1997) SWISS-MODEL and the Swiss-Pdb viewer: an environment for comparative protein modeling. Electrophoresis 18:2714–2723
Gundampati RK, Chikati R, Kumari M, Sharma A, Pratyush DD, Jagannadham MV, Kumar CS, Debnath MD (2012) Protein-protein docking on molecular models of Aspergillus niger RNase and human actin: novel target for anticancer therapeutics. J Mol Model 18(2):653–662
Guruprasad K, Reddy BVB, Pandit MW (1990) Correlation between stability of a protein and its dipeptide composition: a novel approach for predicting in vivo stability of a protein from its primary sequence. Protein Eng 4:155–161
Haberthur U, Caflisch A (2008) FACTS: fast analytical continuum treatment of salvation. J Comput Chem 29:701–715
Hasan M, Hakim A, Iqbal A, Bhuiyan FR, Begum MK, Sharmin S, Abir RA (2015) Computational study and homology modeling of phenol hydroxylase: key enzyme for phenol degradation. Int J Comput Bioinfo In Silico Model 4(4):691–698
Holcenberg JS, Teller DC (1976) Physical properties of antitumor glutaminase-asparaginase from Pseudomonas 7A. J Biol Chem 251:5375–5380
Holcenberg JS, Ericsson L, Roberts J (1978) Amino acid sequence of the diazooxonorleucine binding site of Acinetobacter and Pseudomonas 7 A glutaminase-asparaginase enzymes. Biochemistry 17(3):11–417
Houchens DP, Ovejera AA, Sheridan MA, Johnson RK, Bogden AE, Neil GL (1979) Therapy for mouse tumors and human tumor xenografts with the antitumor antibiotic AT-125. Cancer Treat Rep 63:473–476
Ikai A (1980) Thermostability and aliphatic index of globular proteins. Biochemistry 88:1895–1898
Khobragade CN, Beedkar SD, Bodade RG, Vinchurkar AS (2011) Comparative structural modeling and docking studies of oxalate oxidase: possible implication in enzyme supplementation therapy for urolithiasis. Int J Biol Macromol 48:466–473
Kyte J, Doolittle RF (1982) A simple method for displaying the hydropathic character of a protein. J Mol Biol 157:105–132
Lanka S, Talluri VR, Ganesh V, Latha JNL (2015) Homology modeling, molecular dynamic simulations and docking studies of a new cold active extracellular lipase, EnL A from Emericella nidulans NFCCI 3643. Trends Bioinform 8:37–51
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23(21):2947–2948
Medina MA, Jimenez FS, Marquez J, Quesada AR, Castro IND (1992) Relevance of glutamine metabolism to tumor cell growth. Mol Cell Biochem 113:1–15
Melo F, Feytmans E (1998) Assessing protein structures with non-local atomic interaction energy. J Mol Biol 277:1141–1152
Moharam ME, Gamal-Eldeen AM, El-sayed ST (2010) Production, immobilization and antitumor activity of l-asparaginase of Bacillus sp. R36. J Am Sci 6(8):131–140
Mosmann T (1983) Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods 65:55–63
Nayeem A, Sitkoff D, Krystek S Jr (2006) A comparative study of available software for high-accuracy homology modeling: from sequence alignments to structural models. Protein Sci 15:808–824
Ovejera AA, Houchens DP, Catane R, Sheridan MA, Muggia FM (1979) Efficacy of 6-diazo-5-oxo-l-norleucine and N-[N-gamma-glutamyl-6-diazo-5-oxo-norleucinyl]-6-diazo-5-oxo-norleucine against experimental tumors in conventional and nude mice. Cancer Res 39:3220–3224
Oza VP, Parmar PP, Patel PI, Singh R, Trivedi U, Subramanian RB (2011) Homology modeling of plant l-asparaginase: characterization of its ligand binding efficiency. J Adv Bioinform Appl Res 2(1):100–107
Pace CN, Vajdos F, Fee L, Grimsley G, Gray T (1995) How to measure and predict the molar absorption coefficient of a protein. Protein Sci 11:2411–2423
Pal S, Maity P (1992) Antineoplastic activities of purified bacterial glutaminase on transplanted tumour system. Indian J Cancer Chemother 13:73–76
Poster DS, Bruno S, Penta J, Neil GL, McGovren JP (1981) Acivicin an antitumor antibiotic. Cancer Clin Trials 4:327–330
Reda FM (2015) Kinetic properties of Streptomyces canarius l-glutaminase and its anticancer efficiency. Braz J Microbiol 46(4):957–968
Roberts J, McGregor WG (1991) Inhibition of mouse retroviral disease by bioactive glutaminase-asparaginase. J Gen Virol 72:299–305
Roberts J, McAllister TW, Sethuraman N, Freeman AG (2001) Genetically engineered glutaminase and its use in antiviral and anticancer therapy. US 6312939 B1
Schalk AM, Nguyen HA, Rigouin C, Lavie A (2014) Identification and structural analysis of an l-asparaginase enzyme from guinea pig with putative tumor cell killing properties. J Biol Chem 289(48):33175–33186
Schwede T, Kopp J, Guex N, Peitsch MC (2003) SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res 31:3381–3385
Scotti C, Sommi P, Pasquetto MV, Cappelletti D, Stivala S, Mignosi P, Savio M, Chiarelli LR, Valentini GJJ, Bolanos-Garcia VM, Merrell DS, Franchini S, Verona ML, Bolis C, Solcia E, Manca R, Franciotta D, Casasco A, Filipazzi P, Zardini E, Vannini V (2010) Cell-cycle inhibition by Helicobacter pylori l-asparaginase. PLoS One 5(11):e13892
Senthil KM, Selvam K, Singaravel R (2012) Homology modeling of a unique extracellular glutaminase free l-asparaginase from novel marine actinomycetes. Int J Res Biotechnol Biochem 2(1):1–12
Singh P, Banik RM (2013) Biochemical characterization and antitumor study of l-glutaminase from Bacillus cereus MTCC 1305. Appl Biochem Biotechnol 171:522–531
Sinsuwan S, Yongsawatdigul J, Chumseng S, Yamabhai M (2012) Efficient expression and purification of recombinant glutaminase from Bacillus licheniformis (GlsA) in Escherichia coli. Protein Express Purif 83:52–58
Spiers ASD, Wade HE (1979) Achromobacter l-glutaminase-l-asparaginase: human pharmacology, toxicology, and activity in acute leukemia. Cancer Treat Rep 63:1019–1024
Tobias JW, Shrader TE, Rocap G, Varshavsky A (1991) The N-end rule in bacteria. Science 254:1374–1377
Varshavsky A (1997) The N-end rule pathway of protein degradation. Genes Cells 2:13–28
Vila J, Thomasset N, Navarro C, Dore JF (1990) In vitro and in vivo anti tumor activity of l-glutamic acid gamma-monohydroxamate against L1210 leukemia and B16 melanoma. Int J Cancer 45:737–743
Wang B, Yang LP, Zhang XZ, Huang SQ, Bartlam M, Zhou SF (2009) New insights into the structural characteristics and functional relevance of the human cytochrome P450 2D6 enzyme. Drug Metab Rev 41:573–643
Wu MC, Arimura GK, Holcenberg JS, Yunis AA (1978) Mechanism of sensitivity of cultured pancreatic carcinoma to asparaginase. Int J Cancer 22:728–733
Xiang Z (2006) Advances in homology protein structure modeling. Curr Protein Pept Sci 7:217–227
Yadav M, Singh A, Rathaur S, Liebau E (2010) Structural modeling and simulation studies of Brugia malayi glutathione-S-transferase with compounds exhibiting antifilarial activity: implication in drug targeting and designing. J Mol Graph Model 28:435–445
Yoshimune K, Shirakihara Y, Shiratori A, Wakayama M, Chantawannakul P, Moriguchi M (2006) Crystal structure of a major fragment of the salt-tolerant glutaminase from Micrococcus luteus K-3. Biochem Biophys Res Commun 346:1118–1124
Ziade W, Decombaz AG, Affolter MC (2003) Functional characterization of a salt- and thermotolerant glutaminase from Lactobacillus rhamnosus. Enzyme Microb Technol 32:862–867
Acknowledgments
Authors are thankful to the School of Biochemical Engineering, Indian Institute of Technology (BHU), Varanasi, India for providing research facilities to carry out this work. Mrs. Priyanka Singh is also thankful to DST-INSPIRE for providing financial support in the form of JRF-Professional.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Singh, P., Banik, R.M. & Shah, P. Amino acid sequence determination, in silico tertiary structure prediction and anticancer activity assessment of l-glutaminase from Bacillus cereus . Netw Model Anal Health Inform Bioinforma 5, 11 (2016). https://doi.org/10.1007/s13721-016-0118-5
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13721-016-0118-5