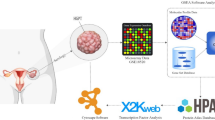
Abstract
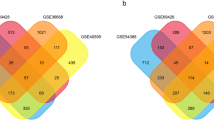
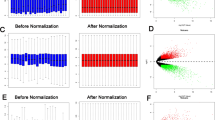
Ovarian cancer (OC) is a frequent fatal malignancy in female reproductive systems with poor early diagnosis. There are several currently utilized popular methods for the detection of OC biomarkers from the microarray transcriptomics dataset, but these methods suffer from the limitation in the identification of biomarkers in the presence of outlier. In this study, we introduced a rule for modification of outlier to improve the performance of biomarker selection methods. We employed the proposed procedure on simulated and three publicly available ovarian cancer gene expression datasets, and improved performance of the proposed procedure was observed. We identified 226 differentially expressed genes (DEGs) overlapped in 3 proposed modified microarray OC datasets using LIMMA in R. These DEGs were underwent Gene Ontology analysis and revealed apoptotic signaling and programmed cell death as an important biological process. The pathway enrichment analysis showed molecular pathways in OC. We also identified FOXC1, GATA2, E2F1, YY1, and FOXL1 as regulatory transcription factors. The protein–protein interaction analysis demonstrated upregulated hub proteins (HDAC1, RPS15, SF3B1, YWHAH, EIF1AX, CALM1, PSME3, UBC, MCL1, NFE2L2), and down-regulated hub proteins (RPL7, RPL9, HSP90AB1, RPS16, RPL30, SKP1, RPL10, RPL14, RPL24, RPLP1). The differential expression of these hub proteins was cross-validated in independent OC RNA-Seq datasets from the TCGA database. The prognostic performance of these hub proteins was observed associated with the worst survival outcomes in OC. Finally, considering 226 DEGs as gene signature, using the L1000CDS2, we revealed 18 drugs in OC with overlap > 0.04. The predicted drugs were repositioned in OC considering the core biomarker signature.










Similar content being viewed by others
References
Aguirre-Gamboa R, Gomez-Rueda H, Martínez-Ledesma E, Martínez-Torteya A, Chacolla-Huaringa R, Rodriguez-Barrientos A, Tamez-Peña JG, Treviño V (2013) SurvExpress: an online biomarker validation tool and database for cancer gene expression data using survival analysis. PLoS ONE 8:1–9. https://doi.org/10.1371/journal.pone.0074250
Altinok G, Powell IJ, Che M, Hormont K, Sarkar FH, Sakr WA, Grignon D, Liao DJ (2006) Reduction of QM protein expression correlates with tumor grade in prostatic adenocarcinoma. Prostate Cancer Prostatic Dis 9:77–82
Asifuzzaman MI, Akter H, Rashid MM, Mollah MNH, Islam SMS, Shahjaman M (2019) Improved k-nearest neighbors approach for incomplete and contaminated gene expression datasets. J Biosci 27:31–41
Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Holko M (2012) NCBI GEO: archive for functional genomics data sets—update. Nucleic Acids Res 41:D991–D995
Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B 57:289–300
Bezerra DP, Pessoa C, de Moraes MO, Saker-Neto N, Silveira ER, Costa-Lotufo LV (2013) Overview of the therapeutic potential of piplartine (piperlongumine). Eur J Pharm Sci 48:453–463
Bharadwaj U, Eckols TK, Kolosov M, Kasembeli MM, Adam A, Torres D, Zhang X, Dobrolecki LE, Wei W, Lewis MT (2015) Drug-repositioning screening identified piperlongumine as a direct STAT3 inhibitor with potent activity against breast cancer. Oncogene 34:1341–1353
Coburn SB, Bray F, Sherman ME, Trabert B (2017) International patterns and trends in ovarian cancer incidence, overall and by histologic subtype. Int J cancer 140:2451–2460
Ferlay J, Shin H, Bray F, Forman D, Mathers C, Parkin DM (2010) Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J cancer 127:2893–2917
Gov E, Kori M, Arga KY (2017) Multiomics analysis of tumor microenvironment reveals Gata2 and miRNA-124-3p as potential novel biomarkers in ovarian cancer. OMICS 21:603–615
Grant S, Easley C, Kirkpatrick P (2007) Vorinostat. Nat Rev Drug Discov 6:21–22. https://doi.org/10.1038/nrd2227
Hayashi A, Horiuchi A, Kikuchi N, Hayashi T, Fuseya C, Suzuki A, Konishi I, Shiozawa T (2010) Type-specific roles of histone deacetylase (HDAC) overexpression in ovarian carcinoma: HDAC1 enhances cell proliferation and HDAC3 stimulates cell migration with downregulation of E-cadherin. Int J cancer 127:1332–1346
Hoque ME, Karim S, Siddiqui MMR, Ahmed T (2017) Report on three cases of advance ovarian cancer upon Bangladeshi population: successful management with bevacizumab based chemotherapy. Anwer Khan Mod Med Coll J 8:157–161
Huang W, Bhavsar A, Ward RE, Hall JCE, Priestley JV, Michael-Titus AT (2009) Arachidonyl trifluoromethyl ketone is neuroprotective after spinal cord injury. J Neurotrauma 26:1429–1434
Islam T, Rahman MR, Shuvo MAH, Shahjaman M, Islam MR, Karim MR (2019) Drug repositioning and biomarkers in low-grade glioma via bioinformatics approach. Informatics Med Unlocked 17:100250
Je EM, Yoo NJ, Kim YJ, Kim MS, Lee SH (2013) Mutational analysis of splicing machinery genes SF3B1, U2AF1 and SRSF2 in myelodysplasia and other common tumors. Int J cancer 133:260–265
Jurczyszyn A, Zebzda A, Czepiel J, Perucki W, Bazan-Socha S, Cibor D, Owczarek D, Majka M (2014) Geldanamycin and its derivatives inhibit the growth of myeloma cells and reduce the expression of the MET receptor. J Cancer 5:480
Kehoe S, Hook J, Nankivell M, Jayson GC, Kitchener H, Lopes T, Luesley D, Perren T, Bannoo S, Mascarenhas M (2015) Primary chemotherapy versus primary surgery for newly diagnosed advanced ovarian cancer (CHORUS): an open-label, randomised, controlled, non-inferiority trial. Lancet 386:249–257
Kruskal WH, Wallis WA (1952) Use of ranks in one-criterion variance analysis. J Am Stat Assoc 47:583–621
Lepage CC, Nachtigal MW, McManus KJ (2019) Diminished SKP1 or CUL1 expression induces chromosome instability in high-grade serous ovarian cancer precursor cells. Am Assoc Cancer Res. https://doi.org/10.1158/1538-7445.AM2019-1739
Matsumura N, Huang Z, Baba T, Lee PS, Barnett JC, Mori S, Chang JT, Kuo W-L, Gusberg AH, Whitaker RS (2009) Yin yang 1 modulates taxane response in epithelial ovarian cancer. Mol Cancer Res 7:210–220
Nadon R, Shoemaker J (2002) Statistical issues with microarrays: processing and analysis. TRENDS Genet 18:265–271
Rahman MR, Islam T, Gov E, Turanli B, Gulfidan G, Shahjaman M, Banu NA, Mollah MNH, Arga KY, Moni MA (2019) Identification of prognostic biomarker signatures and candidate drugs in colorectal cancer: insights from systems biology analysis. Medicina (Kaunas). https://doi.org/10.3390/medicina55010020
Schmid BC, Oehler MK (2014) New perspectives in ovarian cancer treatment. Maturitas 77:128–136
Shahjaman M, Kumar N, Mollah M, Hossain M, Ahmed M, Ara Begum A, Haque N (2017) Robust significance analysis of microarrays by minimum β-divergence method. BioMed Res Int 2017:5310198. https://doi.org/10.1155/2017/5310198
Shahjaman M, Rahman MR, Islam SMS, Mollah MNH (2019) A robust approach for identification of cancer biomarkers and candidate drugs. Medicina (Kaunas). https://doi.org/10.3390/medicina55060269
Shao A, Wang Z, Wu H, Dong X, Li Y, Tu S, Tang J, Zhao M, Zhang J, Hong Y (2016) Enhancement of autophagy by histone deacetylase inhibitor trichostatin a ameliorates neuronal apoptosis after subarachnoid hemorrhage in rats. Mol Neurobiol 53:18–27
Siegel RL, Miller KD, Jemal A (2020) Cancer statistics, 2020. CA Cancer J Clin 70:7–30
Simões-Pereira J, Moura MM, Marques IJ, Rito M, Cabrera RA, Leite V, Cavaco BM (2019) The role of EIF1AX in thyroid cancer tumourigenesis and progression. J Endocrinol Invest 42:313–318
Smyth GK (2004) Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3:1–25. https://doi.org/10.2202/1544-6115.1027
Subramanian A, Narayan R, Corsello SM, Peck DD, Natoli TE, Lu X, Gould J, Davis JF, Tubelli AA, Asiedu JK, Lahr DL, Hirschman JE, Liu Z, Donahue M, Julian B, Khan M, Wadden D, Smith IC, Lam D, Liberzon A, Toder C, Bagul M, Orzechowski M, Enache OM, Piccioni F, Johnson SA, Lyons NJ, Berger AH, Shamji AF, Brooks AN, Vrcic A, Flynn C, Rosains J, Takeda DY, Hu R, Davison D, Lamb J, Ardlie K, Hogstrom L, Greenside P, Gray NS, Clemons PA, Silver S, Wu X, Zhao W-N, Read-Button W, Wu X, Haggarty SJ, Ronco LV, Boehm JS, Schreiber SL, Doench JG, Bittker JA, Root DE, Wong B, Golub TR (2017) A next generation connectivity map: L1000 platform and the first 1,000,000 profiles. Cell 171:1437–1452.e17. https://doi.org/10.1016/j.cell.2017.10.049
Sumanasekera W, Beckmann T, Fuller L, Castle M, Huff M (2018) Epidemiology of ovarian cancer: risk factors and prevention. Biomed J 1:13
Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP (2015) STRING v10: protein–protein interaction networks, integrated over the tree of life. Nucleic Acids Res 43:D447–D452
Torre LA, Trabert B, DeSantis CE, Miller KD, Samimi G, Runowicz CD, Gaudet MM, Jemal A, Siegel RL (2018) Ovarian cancer statistics, 2018. CA Cancer J Clin 68:284–296
Turanli B, Karagoz K, Bidkhori G, Sinha R, Gatza ML, Uhlen M, Mardinoglu A, Arga KY (2019a) Multi-omic data interpretation to repurpose subtype specific drug candidates for breast cancer. Front Genet 10:420. https://doi.org/10.3389/fgene.2019.00420
Turanli B, Zhang C, Kim W, Benfeitas R, Uhlen M, Arga KY, Mardinoglu A (2019b) Discovery of therapeutic agents for prostate cancer using genome-scale metabolic modeling and drug repositioning. EBioMedicine 42:386–396. https://doi.org/10.1016/j.ebiom.2019.03.009
Tusher VG, Tibshirani R, Chu G (2001) Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci 98:5116–5121
Wang B, Liu S-Z, Zheng R-S, Zhang F, Chen W-Q, Sun X-B (2014) Time trends of ovarian cancer incidence in China. Asian Pacific J cancer Prev 15:191–193
Wang L, Li L, Yang Z (2016) Correlation of FOXC1 protein with clinicopathological features in serous ovarian tumors. Oncol Lett 11:933–938
Wilcoxon F (1945) Individual comparisons by ranking methods. Biom Bull 1(6):80–83
Xia J, Gill EE, Hancock REW (2015) NetworkAnalyst for statistical, visual and network-based meta-analysis of gene expression data. Nat Protoc 10:823
Xiang W, Yang C-Y, Bai L (2018) MCL-1 inhibition in cancer treatment. Onco Targets Ther 11:7301
Yu L, Li J-J, Liang X-L, Wu H, Liang Z (2019) PSME3 promotes TGFB1 secretion by pancreatic cancer cells to induce pancreatic stellate cell proliferation. J Cancer 10:2128
Zhan L, Zhang Y, Wang W, Song E, Fan Y, Wei B (2016) E2F1: a promising regulator in ovarian carcinoma. Tumor Biol 37:2823–2831
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
Authors declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Shahjaman, M., Jui, F.T.Z., Islam, T. et al. Improved identification of core biomarkers and drug repositioning for ovarian cancer: an integrated bioinformatics approach. Netw Model Anal Health Inform Bioinforma 9, 62 (2020). https://doi.org/10.1007/s13721-020-00267-2
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13721-020-00267-2