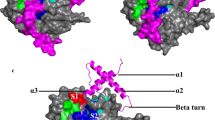
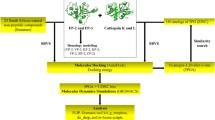
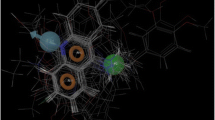
Abstract
Papain-like cysteine protease of P. knowlesi is a significant target for the treatment of malaria infection. P. knowlesi poses a new threat to malaria eradication programs as it starts infecting humans. We have targeted P. knowlesi Knowpain-4 protein for inhibitor designing based on the molecular modeling simulations and structure-based pharmacophore modeling. In-depth sequence analysis reveals its crucial role in host hemoglobin degradation. Dihedral angles and QMEAN score analysis successfully validate the modeled structure of Knowpain4. The docking simulation leads to the prediction of E64 poses at the binding site of Knowpain4 protein. The ligand–receptor interaction information is used for generating five different pharmacophore models taking a diverse set of parameters. A combined 3D features-based, a pharmacophore model is assembled to screen in-house database. The presence of E64, along with 2,128 compounds in the output cross-confirms our screening model. Later on, top 22 screened compounds with higher relative pharmacophore score to E64 is selected for interaction study. The docking simulation experiments result in the identification of 6 compounds that occupies the functional, active site of Knowpain4 protein with high binding affinity. Further, pharmacokinetic studies showed that the identified compounds are potential bioactive molecules with unique scaffolds, and may have the potential to block the expression of Knowpain4 protease.





Similar content being viewed by others
Abbreviations
- ADMET:
-
Absorption, distribution, metabolism, excretion and toxicity
- KP4:
-
Knowpain4
- 3D:
-
Three dimensional
- TPSA:
-
Topological Polar Surface Area
- HA:
-
Hydrogen bond acceptors
- HD:
-
Hydrogen bond donors
- BLAST:
-
Basic Local Alignment Search Tool
- CADD:
-
Computer aided drug design
- NCBI:
-
National Center for Biotechnology Information
- GUI:
-
Graphical user interface
References
Amir A, Russell B, Liew JW, Moon RW, Fong MY, Vythilingam I, Subramaniam V, Snounou G, Lau YL (2016) Invasion characteristics of a Plasmodium knowlesi line newly isolated from a human. Sci Rep 6:24623. https://doi.org/10.1038/srep24623
Bekono BD, Ntie-Kang F, Owono Owono LC, Megnassan E (2018) Targeting cysteine proteases from Plasmodium falciparum: a general overview, rational drug design and computational approaches for drug discovery. Curr Drug Targets 19(5):501–526. https://doi.org/10.2174/1389450117666161221122432
Benkert P, Künzli M, Schwede T (2009) QMEAN server for protein model quality estimation. Nucleic Acids Res 37(Web Server issue):W510–W514. https://doi.org/10.1093/nar/gkp322
Benson DA, Cavanaugh M, Clark K, Karsch-Mizrachi I, Ostell J, Pruitt KD, Sayers EW (2018) GenBank. Nucleic Acids Res 46(D1):D41–D47. https://doi.org/10.1093/nar/gkx1094
Breman JG, Alilio MS, Mills A (2004) Conquering the intolerable burden of malaria: what’s new, what’s needed: a summary. Am J Trop Med Hyg 71:1–15
Burley SK, Berman HM, Kleywegt GJ, Markley JL, Nakamura H, Velankar S (2017) Protein Data Bank (PDB): the single global macromolecular structure archive. Methods Mol Biol 1607:627–641. https://doi.org/10.1007/978-1-4939-7000-1_26
Cox-Singh J, Davis TM, Lee KS, Shamsul SS, Matusop A, Ratnam S, Rahman HA, Conway DJ, Singh B (2008) Plasmodium knowlesi malaria in humans is widely distributed and potentially life threatening. Clin Infect Dis 46(2):165–171. https://doi.org/10.1086/524888
Daneshvar C, Davis TM, Cox-Singh J, Rafa’ee MZ, Zakaria SK, Divis PC, Singh B (2009) Clinical and laboratory features of human Plasmodium knowlesi infection. Clin Infect Dis 49:852–860. https://doi.org/10.1086/605439
Davidson G, Chua TH, Cook A, Speldewinde P, Weinstein P (2019) Defining the ecological and evolutionary drivers of Plasmodium knowlesi transmission within a multi-scale framework. Malar J 18(1):66. https://doi.org/10.1186/s12936-019-2693-2
Kirchmair J, Laggner C, Wolber G, Langer T (2005) Comparative analysis of protein-bound ligand conformations with respect to catalyst’s conformational space subsampling algorithms. J Chem Inf Model 45(2):422–430. https://doi.org/10.1021/ci049753l
Kumar S, Farmer R, Turnbull AP, Tripathy NK, Manjasetty BA (2013) Structural and functional conservation profiles of novel cathepsin L-like proteins identified in the Drosophila melanogaster genome. J Biomol Struct Dyn 31(12):1481–1489. https://doi.org/10.1080/07391102.2012.745379
Lipinski CA (2004) Lead- and drug-like compounds: the rule-of-five revolution. Drug Discov Today Technol 4:337–341. https://doi.org/10.1016/j.ddtec.2004.11.007
Lipinski CA, Lombardo F, Dominy BW, Feeney PJ (2001) Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev 46(1–3):3–26. https://doi.org/10.1016/s0169-409x(00)00129-0
Lovell SC, Davis IW, Arendall WB 3rd, de Bakker PI, Word JM, Prisant MG, Richardson JS, Richardson DC (2003) Structure validation by Calpha geometry: phi, psi and Cbeta deviation. Proteins 50(3):437–450. https://doi.org/10.1002/prot.10286
Loy DE, Liu W, Li Y et al (2017) Out of Africa: origins and evolution of the human malaria parasites Plasmodium falciparum and Plasmodium vivax. Int J Parasitol 47(2–3):87–97. https://doi.org/10.1016/j.ijpara.2016.05.008
Marchler-Bauer A, Derbyshire MK, Gonzales NR et al (2015) CDD: NCBI’s conserved domain database. Nucleic Acids Res 43(Database issue):D222–D226. https://doi.org/10.1093/nar/gku1221
Michael E, Madon S (2017) Socio-ecological dynamics and challenges to the governance of Neglected Tropical Disease control. Infect Dis Poverty 6(1):35. https://doi.org/10.1186/s40249-016-0235-5
Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ (2009) AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem 30(16):2785–2791. https://doi.org/10.1002/jcc.21256
Musyoka TM, Njuguna JN, Tastan Bishop Ö (2019) Comparing sequence and structure of falcipains and human homologs at prodomain and catalytic active site for malarial peptide based inhibitor design. Malar J 18(1):159. https://doi.org/10.1186/s12936-019-2790-2
Na BK, Shenai BR, Sijwali PS, Choe Y, Pandey KC, Singh A, Craik CS, Rosenthal PJ (2004) Identification and biochemical characterization of vivapains, cysteine proteases of the malaria parasite Plasmodium vivax. Biochem J 378:529–538. https://doi.org/10.1042/BJ20031487
Novinec M, Lenarcic B (2013) Papain-like peptidases: structure, function, and evolution. Biomol Concepts 4:287–308. https://doi.org/10.1515/bmc-2012-0054
Pain A, Böhme U, Berry AE et al (2008) The genome of the simian and human malaria parasite Plasmodium knowlesi. Nature 455(7214):799–803. https://doi.org/10.1038/nature07306
Pant A, Kumar R, Wani NA, Verma S, Sharma R, Pande V, Saxena AK, Dixit R, Rai R, Pandey KC (2018) Allosteric site inhibitor disrupting auto-processing of malarial cysteine proteases. Sci Rep 8(1):16193. https://doi.org/10.1038/s41598-018-34564-8
Pasini EM, Zeeman AM, Voorberg-van der Wel A, Kocken CHM (2018) Plasmodium knowlesi: a relevant, versatile experimental malaria model. Parasitology 145(1):56–70. https://doi.org/10.1017/S0031182016002286
Prasad R, Soni A, Puri SK, Sijwali PS (2012) Expression, characterization, and cellular localization of knowpains, papain-like cysteine proteases of the Plasmodium knowlesi malaria parasite. PLoS ONE 7:e51619. https://doi.org/10.1371/journal.pone.0051619
Rosenthal PJ (2020) Falcipain cysteine proteases of malaria parasites: an update. Biochim Biophys Acta Proteins Proteom 1868(3):140362. https://doi.org/10.1016/j.bbapap.2020.140362
Sterling T, Irwin JJ (2015) ZINC 15–ligand discovery for everyone. J Chem Inf Model 55(11):2324–2337. https://doi.org/10.1021/acs.jcim.5b00559
Trott O, Olson AJ (2010) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 31(2):455–461. https://doi.org/10.1002/jcc.21334
Webb B, Sali A (2017) Protein structure modeling with MODELLER. Methods Mol Biol 1654:39–54. https://doi.org/10.1007/978-1-4939-7231-9_4
William T, Rahman HA, Jelip J, Ibrahim MY, Menon J, Grigg MJ, Yeo TW, Anstey NM, Barber BE (2013) Increasing incidence of Plasmodium knowlesi malaria following control of P. falciparum and P. vivax Malaria in Sabah, Malaysia. PLoS Negl Trop Dis 7(1):e2026. https://doi.org/10.1371/journal.pntd.0002026
Wishart DS, Feunang YD, Guo AC et al (2018) DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res 46(D1):D1074–D1082. https://doi.org/10.1093/nar/gkx1037
Wolber G, Langer T (2005) LigandScout: 3-D pharmacophores derived from protein-bound ligands and their use as virtual screening filters. J Chem Inf Model 45(1):160–169. https://doi.org/10.1021/ci049885e
Yadav MK, Swati D (2012) Comparative genome analysis of six malarial parasites using codon usage bias based tools. Bioinformation 8:1230–1239. https://doi.org/10.6026/97320630081230
Yadav MK, Singh A, Swati D (2014) A knowledge-based approach for identification of drugs against vivapain-2 protein of Plasmodium vivax through pharmacophore-based virtual screening with comparative modelling. Appl Biochem Biotechnol 173:2174–2188. https://doi.org/10.1007/s12010-014-1023-y
Yang J, Roy A, Zhang Y (2013) Protein-ligand binding site recognition using complementary binding-specific substructure comparison and sequence profile alignment. Bioinformatics 29(20):2588–2595. https://doi.org/10.1093/bioinformatics/btt447
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Yadav, M.K., Bharti, R.R. & Rashmi, M. Exploration and identification of novel inhibitors against Knowpain-4 of P. knowlesi using a combinatorial 3D pharmacophore modeling approach. Netw Model Anal Health Inform Bioinforma 9, 67 (2020). https://doi.org/10.1007/s13721-020-00273-4
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13721-020-00273-4