Abstract
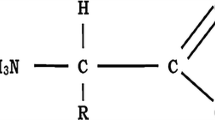
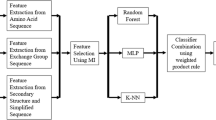
Proteins form the basis of all major life processes that sustain life. The functionality of a protein is a direct consequence of its underlying structure. Protein structure prediction thus serves to ascertain the function of similar or dissimilar proteins, accordingly. Secondary structure prediction paves way for 3D structures that eventually decides protein properties. It also aims to facilitate probable structures for proteins whose structures remain undiscovered. Although experimental approaches have been quite efficient in extracting protein secondary structure from its amino acid sequence, yet it is often cumbersome and time intensive to achieve it in vitro. Hence, computational approaches are required to predict secondary structures for the diverse amino acids constituting these proteins. However, the available computational models fail to register good prediction accuracy due to inadequate modelling of sequence-structure relationship. Also, the dearth of global exploration-based methods further makes them ineffective in catering to the evolving proteomic data. Accordingly, PSO (Particle swarm optimization) has been explored to propose a neural network model for protein secondary structure prediction (PSSP). Six standard datasets namely- PSS504, RS126, EVA6, CB396, Manesh and CB513 have been utilized for the training and testing of the neural network. The proposed model is evaluated on the basis of its Q3 accuracy, precision, and recall. The 10, 20, 30 and 40 fold cross validation in combination with sensitivity analysis and has been carried out for verification of results. The proposed model is found to outperform most of the existing models by demonstrating a better average Q3 accuracy lying above 81% for PSSP.





Similar content being viewed by others
References
Ahmed H, Glasgow J (2012) Swarm intelligence: concepts, models and applications, school of computing. Queen’s University Kingston, Ontario
Alberts B, Bray D, Johnson A, Lewis J, Raff M, Roberts K, Walter P (2004) Essential cell biology. 2nd (ed.). Garland Science
AlGhamdi R, Aziz A, Alshehri M et al (2020) Deep learning model with ensemble techniques to compute the secondary structure of proteins. J Supercomput. https://doi.org/10.1007/s11227-020-03467-9
Altschul S, Gish W, Miller W, Myers E, Lipman D (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Bishop C (1995) Neural networks for pattern recognition. Oxford University Press, Oxford
Branden C, Tooze J (1999) Introduction to protein structure. Garland Pub, New York
Busia A (2017) Next-step conditioned deep convolutional neural networks improve protein secondary structure prediction. arXiv:1702.03865v1. Accessed date 13 Feb 2017
Chandonia J, Karplus M (1995) Neural networks for secondary structure and structural class predictions. Protein Sci 4:275–285
Chang DT et al (2008) Prediction of protein secondary structures with a novel kernel density estimation based classifier. BMC Res Notes 1(1):51
Chen K, Kurgan L, Ruan J (2006) Optimization of the sliding window size for protein structure prediction. 2006 IEEE symposium on computational intelligence and bioinformatics and computational biology, Toronto, ON, Canada, pp. 1–7. https://doi.org/10.1109/CIBCB.2006.330959
Chin YF et al. (2011) Optimized local protein structure with support vector machine to predict protein secondary structure. Proceedings of the 3rd Malaysian joint conference on artificial lntelligence, July 2011
Chou PY, Fasman UD (1974) Prediction of protein conformation. Biochemistry 13:211–215
Cuff JA, Barton GJ (1999) Evaluation and improvement of multiple sequence methods for protein secondary structure prediction. Proteins Struct Funct Bioinf 34(4):508–519
Custódio FL et al (2014) A multiple minima genetic algorithm for protein structure prediction. Appl Soft Comput 15:88–99
Cutello V et al (2005) A multi-objective evolutionary approach to the protein structure prediction problem. J R Soc Interface 3(6):139–151
Das S, Abraham A, Konar A (2008) Swarm intelligence algorithms in bioinformatics studies. Computational intelligence, vol 94. Springer, Berlin, pp 113–147
Finkelstein AV, Ptitsyn OB (1971) Statistical analysis of the correlation among amino acid residues in helical, βstructural and non-regular regions of globular proteins. J Mol Biol 62:613–624
Gao P et al (2017) A database assisted protein structure prediction method via a swarm intelligence algorithm. R Soc Chem Adv 7(63):39869–39876
Garnier J, Gibrat JF, Robson B (1996) GOR method for predicting protein secondary structure from amino acid sequence. Methods Enzymol 266:540–553
Garro BA et al. (2015) Designing artificial neural networks using particle swarm optimization algorithms. Comput Intell Neurosci
Gubbi J et al (2006) Protein secondary structure prediction using support vector machines and a new feature representation. Int J Comput Intell Appl 551:1
Guo Y et al (2019) DeepACLSTM: deep asymmetric convolutional long short-term memory neural models for protein secondary structure prediction. BMC Bioinf 20(1):1–2
Hao W et al. (2011) A fuzzy adaptive particle swarm optimization for rna secondarystructure prediction. International conference on information science and technology, Nanjing, Jiangsu, China
Jiang Q (2017) Protein secondary structure prediction: a survey of the state of the art. J Mol Graph Model 76:379–402
Karami Y et al. (2012) Protein structure prediction using bio-inspired algorithm: a review. The 16th CSI international symposium on artificial intelligence and signal processing (AISP 2012)
Kennedy J, Eberhart RC (1995) Particle swarm optimization. In: Proceedings of IEEE international conference on neural networks, Perth, Australia, pp. 1942–1948
Kneller DG, Cohen FE, Langridge R (1990) Improvements in protein secondary structure prediction by an enhanced neural network. J Mol Biol 214:171–182
Koswatta TJ, Samaraweera P, Sumanasinghe VA (2011) A simple comparison between specific protein secondary structure prediction tools. Postgraduate Institute of Agriculture University of Peradeniya, Sri Lanka
Lee L et al. (2009) Protein secondary structure predictionusing rule induction from coverings. IEEE conference on computational intelligence in bioinformatics and computational biology, IEEE Press, 2009
Li R et al. (2016) Deep convolutional neural networks for detecting secondary structures in protein density maps from cryo-electron microscopy. IEEE international conference on bioinformatics and biomedicine (BIBM)
Lim VI (1974) Structural principles of the globular organization of protein chains. A stereochemical theory of globular protein secondary structure. J Mol Biol 88:857–872
Lim CP, Jain LC, Dehuri S (2009) Innovations in swarm intelligence: studies in computational intelligence, vol 248. Springer, Berlin
Lin K, Simossis VA, Taylor WR (2005) A simple and fast secondary structure prediction method using hidden neural networks. Bioinformatics 21(2):152–159
Liu W et al. (2006) A hybrid particle swarm optimization algorithm forpredicting the chaotic time series. Proceedings of the 2006 IEEE international conference on mechatronics and automation, Luoyang, China, 2006
Maclin R, Shavlik JW (1993) Using knowledge-based neural networks to improve algorithms: refining the ChouFasman algorithm for protein folding. Mach Learn 11:195–215
Magnan CN et al (2014) SSpro/ACCpro 5: almost perfect prediction of protein secondary structure and relative solvent accessibility using profiles, machine learning and structural similarity. Bioinformatics 30(18):2592–2597
Makolo AU et al (2018) Protein secondary structure prediction using deep neural network and particle swarm optimization algorithm. Int J Comput Appl 181(28):0975–8887
Mansour N et al (2012) Particle swarm optimization approach for protein structure prediction in the 3D HP model. Interdiscip Sci Comput Life Sci 4(3):190–200
McGuffin LJ, Bryson K, Jones DT (2000) The PSIPRED protein structure prediction server. Bioinformatics 16(4):404–405
Naderi-Manesh H, Sadeghi M, Araf S, Movahedi AAM (2001) Predicting of protein surface accessibility with information theory. Proteins 42:452–459
Nakai K, Kidera A, Kanehisa M (1988) Cluster analysis of amino acid indices for prediction of protein structure and function. Protein Eng 2:93–100
Pacifico LDS et al. (2012) Improved group search optimization based onopposite populations for feedforward networkstraining with weight decay. IEEE international conference on systems, man, and cybernetics, COEX, Seoul, Korea
Parsopoulos KE, Vrahatis MN (2010) Particle swarm optimization and intelligence: advances and applications: information science reference. Hershey, Pennsylvania
Pearson K (1904) Mathematical contribution to the theory of evolution. Biometrika 3:131–1390
Qu W et al (2011) Improving protein secondary structure prediction using a multi-modal BP method. Comp Biol Med 41(10):946
Rose GD (1978) Prediction of chain turns in globular proteins on a hydrophobic basis. Nature 272:586–590
Rost B (1996) PHD: predicting one-dimensional protein structure by profile based neural networks. Methods Enzymol 266:525–539
Rost B, Eyrich VA (2001) EVA: large-scale analysis of secondary structure prediction. Proteins Struct Funct Bioinf 45(S5):192–199
Rost B, Sander C, Casadio R, Fariselli P (1995) Transmembrane helices predicted at 95% accuracy. Protein Sci 4(3):521–533
Sen T, Jernigan R, Garnier J, Kloczkowski A (2005) GOR V server for protein secondary structure prediction. Bioinformatics 21(11):2787–2788
Stolorz P, Lapedes A, Xia Y (1992) Predicting protein secondary structure using neural net and statistical methods. J Mol Biol 225:363–377
Wang Y (2017) Protein secondary structure prediction by using deep learning method. Knowl Based Syst 118:115–123
Wang S, Peng J, Ma J et al (2016) Protein secondary structure prediction using deep convolutional neural fields. Sci Rep 6(1):1–1
Wardah W et al (2019) Protein secondary structure prediction using neural networks and deep learning: a review. Comput Biol Chem 81:1
Yang W et al (2013) Prediction of protein secondary structure using Large Margin Nearest Neighbour classification. Int J Bioinf Res Appl 9(2):207
Yao X et al (1997) A new evolutionary system for evolving artificial neural networks. IEEE Trans Neural Netw 8(3):1
Yüksektepe FÜ et al (2008) Prediction of secondary structures of proteins using a two-stage method. Comput Chem Eng 32(1–2):1
Zhang Y et al. (2013) Swarm intelligence and its applications. Sci World J
Zhang J et al (2015) Prediction of protein solvent accessibility using PSO-SVR with multiple sequence-derived features and weighted sliding window scheme. BioData Min 8(1):1–5
Zheng W et al. (2016) Protein secondary structure prediction via Pigeon-Inspired Optimization. 2016 IEEE Chinese guidance, navigation and control conference (CGNCC)
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Akbar, S., Pardasani, K.R. & Khan, F. Swarm optimization-based neural network model for secondary structure prediction of proteins. Netw Model Anal Health Inform Bioinforma 10, 33 (2021). https://doi.org/10.1007/s13721-021-00304-8
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13721-021-00304-8