Abstract
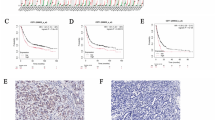
Chromodomain helicase DNA (CHD) binding protein is one class of chromatin remodeling factors, which are essential in gastric cancer (GC) development. However, the role of distinct CHD family member in GC is rarely understood. To resolve this issue, a comprehensive analysis of the prognostic significance of CHD members in GC was performed using Oncomine, UALCAN, GEPIA, Kaplan–Meier Plotter, cBioPortal, LinkedOmics and STRING databases. The mRNA expression of all CHD members was different in GC samples compared with normal controls, and CHD 1/2/3/6/9 expression was remarkedly correlated with advanced cancer stage. Higher CHD3/4/6/8 expression was found significantly associated with poor overall survival (OS), post-progression survival (PPS) and progression-free survival (PFS). Conversely, higher CHD5 expression was remarkedly correlated with better OS, PPS, PFS. The study reveals that CHD3/4/5/6/8 could serve as prognostic biomarkers for GC patients. However, future researches are required to assess the clinical utility of CHD members in GC.







Similar content being viewed by others
References
Badodi S et al (2017) Convergence of BMI1 and CHD7 on ERK signaling in medulloblastoma. Cell Rep 21:2772–2784. https://doi.org/10.1016/j.celrep.2017.11.021
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A (2018) Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 68:394–424. https://doi.org/10.3322/caac.21492
Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi B, Varambally S (2017) UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia 19:649–658. https://doi.org/10.1016/j.neo.2017.05.002
Chu X, Guo X, Jiang Y, Yu H, Liu L, Shan W, Yang ZQ (2017) Genotranscriptomic meta-analysis of the CHD family chromatin remodelers in human cancers—initial evidence of an oncogenic role for CHD7. Mol Oncol 11:1348–1360. https://doi.org/10.1002/1878-0261.12104
Clapier CR, Cairns BR (2009) The biology of chromatin remodeling complexes. Annu Rev Biochem 78:273–304. https://doi.org/10.1146/annurev.biochem.77.062706.153223
Cristescu R et al (2015) Molecular analysis of gastric cancer identifies subtypes associated with distinct clinical outcomes. Nat Med 21:449–456. https://doi.org/10.1038/nm.3850
D’Alesio C et al (2016) RNAi screens identify CHD4 as an essential gene in breast cancer growth. Oncotarget 7:80901–80915. https://doi.org/10.18632/oncotarget.12646
D’Alesio C et al (2019) The chromodomain helicase CHD4 regulates ERBB2 signaling pathway and autophagy in ERBB2(+) breast cancer cells. Biol Open. https://doi.org/10.1242/bio.038323
Gao J et al (2013) Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 6:pl1. https://doi.org/10.1126/scisignal.2004088
Guan B, Ran XG, Du YQ, Ren F, Tian Y, Wang Y, Chen MM (2018) High CHD9 expression is associated with poor prognosis in clear cell renal cell carcinoma. Int J Clin Exp Pathol 11:3697–3702
Gui Y et al (2011) Frequent mutations of chromatin remodeling genes in transitional cell carcinoma of the bladder. Nat Genet 43:875–878. https://doi.org/10.1038/ng.907
Hall JA, Georgel PT (2007) CHD proteins: a diverse family with strong ties. Biochem Cell Biol 85:463–476. https://doi.org/10.1139/o07-063
Hashimoto T et al (2020) Clinical significance of chromatin remodeling factor CHD5 expression in gastric cancer. Oncol Lett 19:1066–1073. https://doi.org/10.3892/ol.2019.11138
Kim MS, Chung NG, Kang MR, Yoo NJ, Lee SH (2011) Genetic and expressional alterations of CHD genes in gastric and colorectal cancers. Histopathology 58:660–668. https://doi.org/10.1111/j.1365-2559.2011.03819.x
Marfella CG, Imbalzano AN (2007) The Chd family of chromatin remodelers. Mutat Res 618:30–40. https://doi.org/10.1016/j.mrfmmm.2006.07.012
McKenzie LD et al (2019) CHD4 regulates the DNA damage response and RAD51 expression in glioblastoma. Sci Rep 9:4444. https://doi.org/10.1038/s41598-019-40327-w
Mills AA (2017) The chromodomain helicase DNA-binding chromatin remodelers: family traits that protect from and promote cancer. Cold Spring Harb Perspect Med. https://doi.org/10.1101/cshperspect.a026450
Mouradov D et al (2014) Colorectal cancer cell lines are representative models of the main molecular subtypes of primary cancer. Cancer Res 74:3238–3247. https://doi.org/10.1158/0008-5472.Can-14-0013
Nagarajan P, Onami TM, Rajagopalan S, Kania S, Donnell R, Venkatachalam S (2009) Role of chromodomain helicase DNA-binding protein 2 in DNA damage response signaling and tumorigenesis. Oncogene 28:1053–1062. https://doi.org/10.1038/onc.2008.440
Nickerson ML et al (2017) Molecular analysis of urothelial cancer cell lines for modeling tumor biology and drug response. Oncogene 36:35–46. https://doi.org/10.1038/onc.2016.172
Nio K et al (2015) Defeating EpCAM(+) liver cancer stem cells by targeting chromatin remodeling enzyme CHD4 in human hepatocellular carcinoma. J Hepatol 63:1164–1172. https://doi.org/10.1016/j.jhep.2015.06.009
Rhodes DR et al (2004) ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia 6:1–6. https://doi.org/10.1016/s1476-5586(04)80047-2
Rodríguez D et al (2015) Mutations in CHD2 cause defective association with active chromatin in chronic lymphocytic leukemia. Blood 126:195–202. https://doi.org/10.1182/blood-2014-10-604959
Sawada G et al (2013) CHD8 is an independent prognostic indicator that regulates Wnt/β-catenin signaling and the cell cycle in gastric cancer. Oncol Rep 30:1137–1142. https://doi.org/10.3892/or.2013.2597
Szász AM et al (2016) Cross-validation of survival associated biomarkers in gastric cancer using transcriptomic data of 1,065 patients. Oncotarget 7:49322–49333. https://doi.org/10.18632/oncotarget.10337
Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z (2017) GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res 45:W98-w102. https://doi.org/10.1093/nar/gkx247
Valencia AM, Kadoch C (2019) Chromatin regulatory mechanisms and therapeutic opportunities in cancer. Nat Cell Biol 21:152–161. https://doi.org/10.1038/s41556-018-0258-1
Van Cutsem E et al (2011) The diagnosis and management of gastric cancer: expert discussion and recommendations from the 12th ESMO/World Congress on Gastrointestinal Cancer, Barcelona, 2010. Ann Oncol 22(Suppl 5):v1–v9. https://doi.org/10.1093/annonc/mdr284
Vasaikar SV, Straub P, Wang J, Zhang B (2018) LinkedOmics: analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res 46:D956-d963. https://doi.org/10.1093/nar/gkx1090
von Mering C, Huynen M, Jaeggi D, Schmidt S, Bork P, Snel B (2003) STRING: a database of predicted functional associations between proteins. Nucleic Acids Res 31:258–261. https://doi.org/10.1093/nar/gkg034
Wang X, Lau KK, So LK, Lam YW (2009) CHD5 is down-regulated through promoter hypermethylation in gastric cancer. J Biomed Sci 16:95. https://doi.org/10.1186/1423-0127-16-95
Xu G, Zhu H, Zhang M, Xu J (2018a) Histone deacetylase 3 is associated with gastric cancer cell growth via the miR-454-mediated targeting of CHD5. Int J Mol Med 41:155–163. https://doi.org/10.3892/ijmm.2017.3225
Xu L, Peng H, Huang XX, Xia YB, Hu KF, Zhang ZM (2018b) Decreased expression of chromodomain helicase DNA-binding protein 9 is a novel independent prognostic biomarker for colorectal cancer. Braz J Med Biol Res 51:e7588. https://doi.org/10.1590/1414-431x20187588
Xu N et al (2020) CHD4 mediates proliferation and migration of non-small cell lung cancer via the RhoA/ROCK pathway by regulating PHF5A. BMC Cancer 20:262. https://doi.org/10.1186/s12885-020-06762-z
Yu H, Jiang Y, Liu L, Shan W, Chu X, Yang Z, Yang ZQ (2017) Integrative genomic and transcriptomic analysis for pinpointing recurrent alterations of plant homeodomain genes and their clinical significance in breast cancer. Oncotarget 8:13099–13115. https://doi.org/10.18632/oncotarget.14402
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Author information
Authors and Affiliations
Contributions
QZ and LX designed and carried out this study; LX analyzed the data and wrote this paper; QZ finally provided language help.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Xu, L., Zheng, Q. CHD3/4/5/6/8 serve as potential prognostic predictors in human gastric cancer. Netw Model Anal Health Inform Bioinforma 10, 34 (2021). https://doi.org/10.1007/s13721-021-00307-5
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13721-021-00307-5