Abstract
Background
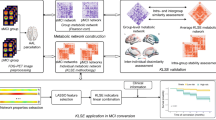
Radiomics-based morphological brain networks (radMBN) constructed from routinely acquired structural MRI (sMRI) data have gained attention in Alzheimer's disease (AD). However, the radMBN suffers from limited characterization of AD because sMRI only characterizes anatomical changes and is not a direct measure of neuronal pathology or brain activity.
Purpose
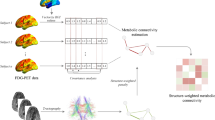
To establish a group sparse representation of the radMBN under a joint constraint of group-level white matter fiber connectivity and individual-level sMRI regional similarity (JCGS-radMBN).
Methods
Two publicly available datasets were adopted, including 120 subjects from ADNI with both T1-weighted image (T1WI) and diffusion MRI (dMRI) for JCGS-radMBN construction, 818 subjects from ADNI and 200 subjects solely with T1WI from AIBL for validation in early AD diagnosis. Specifically, the JCGS-radMBN was conducted by jointly estimating non-zero connections among subjects, with the regularization term constrained by group-level white matter fiber connectivity and individual-level sMRI regional similarity. Then, a triplet graph convolutional network was adopted for early AD diagnosis. The discriminative brain connections were identified using a two-sample t-test, and the neurobiological interpretation was validated by correlating the discriminative brain connections with cognitive scores.
Results
The JCGS-radMBN exhibited superior classification performance over five brain network construction methods. For the typical NC vs. AD classification, the JCGS-radMBN increased by 1–30% in accuracy over the alternatives on ADNI and AIBL. The discriminative brain connections exhibited a strong connectivity to hippocampus, parahippocampal gyrus, and basal ganglia, and had significant correlation with MMSE scores.
Conclusion
The proposed JCGS-radMBN facilitated the AD characterization of brain network established on routinely acquired imaging modality of sMRI.




Similar content being viewed by others
Data availability
The data utilized in this study were sourced from the Alzheimer's Disease Neuroimaging Initiative (ADNI), available at http://adni.loni.usc.edu/data-samples/access-data, and the Australian Imaging, Biomarkers and Lifestyle Flagship Study of Ageing (AIBL), available at http://aibl.csiro.au.
References
Liu J, Li M, Pan Y, Lan W, Zheng R, Wu F-X, et al. Complex brain network analysis and its applications to brain disorders: a survey. Complexity. 2017;2017:8362741.
Yao Z, Hu B, Xie Y, Moore P, Zheng J. A review of structural and functional brain networks: small world and atlas. Brain Inform. 2015;2:45–52.
Supekar K, Menon V, Rubin D, Musen M, Greicius MD. Network analysis of intrinsic functional brain connectivity in Alzheimer’s disease. PLoS Comput Biol. 2008;4(6): e1000100.
Zuo N, Cheng J, Jiang T. Diffusion magnetic resonance imaging for Brainnetome: a critical review. Neurosci Bull. 2012;28:375–88.
Feng J, Zhang S-W, Chen L, Zuo C, AsDN Initiative. Detection of Alzheimer’s disease using features of brain region-of-interest-based individual network constructed with the sMRI image. Comput Med Imaging Graph. 2022;98:102057.
He Y, Chen ZJ, Evans AC. Small-world anatomical networks in the human brain revealed by cortical thickness from MRI. Cereb Cortex. 2007;17(10):2407–19.
He Y, Chen Z, Evans A. Structural insights into aberrant topological patterns of large-scale cortical networks in Alzheimer’s disease. J Neurosci. 2008;28(18):4756–66.
Yao Z, Zhang Y, Lin L, Zhou Y, Xu C, Jiang T, et al. Abnormal cortical networks in mild cognitive impairment and Alzheimer’s disease. PLoS Comput Biol. 2010;6(11): e1001006.
Frisoni GB, Fox NC, Jack CR Jr, Scheltens P, Thompson PM. The clinical use of structural MRI in Alzheimer disease. Nat Rev Neurol. 2010;6(2):67–77.
Subaramya S, Kokul T, Nagulan R, Pinidiyaarachchi U, Jeyasuthan M. Detection of Alzheimer’s disease using structural brain network and convolutional neural network. In: 2021 10th International conference on information and automation for sustainability (ICIAfS), 2021. IEEE; 2021. p. 173–8.
Vemuri P, Jack C. Role of structural MRI in Alzheimer’s disease. Alzheimer’s Res Ther. 2010;2(4):1–10.
Zhao K, Zheng Q, Che T, Dyrba M, Li Q, Ding Y, et al. Regional radiomics similarity networks (R2SNs) in the human brain: reproducibility, small-world properties and a biological basis. Netw Neurosci. 2021;5(3):783–97.
Zhao K, Zheng Q, Dyrba M, Rittman T, Li A, Che T, et al. Regional radiomics similarity networks reveal distinct subtypes and abnormality patterns in mild cognitive impairment. Adv Sci. 2022;9(12): e2104538.
Staffaroni AM, Elahi FM, McDermott D, Marton K, Karageorgiou E, Sacco S, et al. Neuroimaging in dementia. Semin Neurol. 2017;37(05):510–37.
Ghanbari M, Soussia M, Jiang W, Wei D, Yap P-T, Shen D, et al. Alterations of dynamic redundancy of functional brain subnetworks in Alzheimer’s disease and major depression disorders. NeuroImage Clin. 2022;33: 102917.
Ghanbari M, Li G, Hsu LM, Yap PT. Accumulation of network redundancy marks the early stage of Alzheimer’s disease. Hum Brain Mapp. 2023;44(8):2993–3006.
Li Y, Zhang X, Nie J, Zhang G, Fang R, Xu X, et al. Brain connectivity based graph convolutional networks and its application to infant age prediction. IEEE Trans Med Imaging. 2022;41(10):2764–76.
Jin S, Zeng X, Xia F, Huang W, Liu X. Application of deep learning methods in biological networks. Brief Bioinform. 2021;22(2):1902–17.
Zhang X-M, Liang L, Liu L, Tang M-J. Graph neural networks and their current applications in bioinformatics. Front Genet. 2021;12: 690049.
Zhou J, Cui G, Hu S, Zhang Z, Yang C, Liu Z, et al. Graph neural networks: a review of methods and applications. AI Open. 2020;1:57–81.
Zhao X, Wu J, Peng H, Beheshti A, Monaghan JJ, McAlpine D, et al. Deep reinforcement learning guided graph neural networks for brain network analysis. Neural Netw. 2022;154:56–67.
Parisot S, Ktena SI, Ferrante E, Lee M, Guerrero R, Glocker B, et al. Disease prediction using graph convolutional networks: application to autism spectrum disorder and Alzheimer’s disease. Med Image Anal. 2018;48:117–30.
Lee H, Lee DS, Kang H, Kim B-N, Chung MK. Sparse brain network recovery under compressed sensing. IEEE Trans Med Imaging. 2011;30(5):1154–65.
Rosa MJ, Portugal L, Hahn T, Fallgatter AJ, Garrido MI, Shawe-Taylor J, et al. Sparse network-based models for patient classification using fMRI. Neuroimage. 2015;105:493–506.
Yu R, Zhang H, An L, Chen X, Wei Z, Shen D. Connectivity strength-weighted sparse group representation-based brain network construction for MCI classification. Hum Brain Mapp. 2017;38(5):2370–83.
Wee C-Y, Yap P-T, Zhang D, Wang L, Shen D. Group-constrained sparse fMRI connectivity modeling for mild cognitive impairment identification. Brain Struct Funct. 2014;219:641–56.
Song X, Zhou F, Frangi AF, Cao J, Xiao X, Lei Y, et al. Multicenter and multichannel pooling GCN for early AD diagnosis based on dual-modality fused brain network. IEEE Trans Med Imaging. 2022;42(2):354–67.
Zhu D, Zhang T, Jiang X, Hu X, Chen H, Yang N, et al. Fusing DTI and fMRI data: a survey of methods and applications. Neuroimage. 2014;102:184–91.
Hoffer E, Ailon N. Deep metric learning using triplet network. In: Similarity-based pattern recognition: third international workshop, SIMBAD 2015, Copenhagen, Denmark, 12–14 October 2015, Proceedings 3. Springer; 2015. p. 84–92.
Schroff F, Kalenichenko D, Philbin J. FaceNet: a unified embedding for face recognition and clustering. In: Proceedings of the IEEE conference on computer vision and pattern recognition, 2015. p. 815–23.
Wright J, Yang AY, Ganesh A, Sastry SS, Ma Y. Robust face recognition via sparse representation. IEEE Trans Pattern Anal Mach Intell. 2008;31(2):210–27.
Wu Z, Pan S, Chen F, Long G, Zhang C, Philip SY, et al. A comprehensive survey on graph neural networks. IEEE Trans Neural Netw. 2020;32(1):4–24.
Cui Z, Zhong S, Xu P, He Y, Gong G. PANDA: a pipeline toolbox for analyzing brain diffusion images. Front Hum Neurosci. 2013;7:42.
Simpson SL, Bowman FD, Laurienti PJ. Analyzing complex functional brain networks: fusing statistics and network science to understand the brain. Stat Surv. 2013;7:1.
Bullmore E, Sporns O. Complex brain networks: graph theoretical analysis of structural and functional systems. Nat Rev Neurosci. 2009;10(3):186–98.
Dhifallah S, Rekik I, AsDN Initiative. Estimation of connectional brain templates using selective multi-view network normalization. Med Image Anal. 2020;59:101567.
Liu J, Ji S, Ye J. SLEP: sparse learning with efficient projections. Arizona State University; 2009.
Zhang Y, Zhang H, Chen X, Liu M, Zhu X, Lee S-W, et al. Strength and similarity guided group-level brain functional network construction for MCI diagnosis. Pattern Recognit. 2019;88:421–30.
Sampathkumar VR. Adiag: graph neural network based diagnosis of Alzheimer's disease. https://doi.org/10.48550/arXiv.2101.02870.
Fan C-C, Yang H, Peng L, Zhou X-H, Ni Z-L, Zhou Y-J, et al. BGL-Net: a brain-inspired global–local information fusion network for Alzheimer’s disease based on sMRI. IEEE Trans Cogn Dev Syst. 2023;15(3):1161–9.
Wee C-Y, Liu C, Lee A, Poh JS, Ji H, Qiu A, et al. Cortical graph neural network for AD and MCI diagnosis and transfer learning across populations. NeuroImage Clin. 2019;23: 101929.
Wei Y, Price SJ, Schönlieb C-B, Li C. Predicting conversion of mild cognitive impairment to Alzheimer's disease. 2022. https://doi.org/10.48550/arXiv.2203.04725.
Tian X, Liu Y, Wang L, Zeng X, Huang Y, Wang Z. An extensible hierarchical graph convolutional network for early Alzheimer’s disease identification. Comput Methods Programs Biomed. 2023;238: 107597.
Wig GS, Schlaggar BL, Petersen SE. Concepts and principles in the analysis of brain networks. Ann NY Acad Sci. 2011;1224(1):126–46.
Tijms BM, Wink AM, de Haan W, van der Flier WM, Stam CJ, Scheltens P, et al. Alzheimer’s disease: connecting findings from graph theoretical studies of brain networks. Neurobiol Aging. 2013;34(8):2023–36.
Braak H, Braak E, Bohl J. Staging of Alzheimer-related cortical destruction. Eur Neurol. 1993;33(6):403–8.
Fox N, Warrington E, Freeborough P, Hartikainen P, Kennedy A, Stevens J, et al. Presymptomatic hippocampal atrophy in Alzheimer’s disease: a longitudinal MRI study. Brain. 1996;119(6):2001–7.
Köhler S, Black S, Sinden M, Szekely C, Kidron D, Parker J, et al. Memory impairments associated with hippocampal versus parahippocampal-gyrus atrophy: an MR volumetry study in Alzheimer’s disease. Neuropsychologia. 1998;36(9):901–14.
Xue J, Guo H, Gao Y, Wang X, Cui H, Chen Z, et al. Altered directed functional connectivity of the hippocampus in mild cognitive impairment and Alzheimer’s disease: a resting-state fMRI study. Front Aging Neurosci. 2019;11:326.
Seo EH, Lee DY, Lee J-M, Park J-S, Sohn BK, Lee DS, et al. Whole-brain functional networks in cognitively normal, mild cognitive impairment, and Alzheimer’s disease. PLoS ONE. 2013;8(1): e53922.
Miao D, Zhou X, Wu X, Chen C, Tian L. Hippocampal morphological atrophy and distinct patterns of structural covariance network in Alzheimer’s disease and mild cognitive impairment. Front Psychol. 2022;13: 980954.
Feng F, Huang W, Meng Q, Hao W, Yao H, Zhou B, et al. Altered volume and structural connectivity of the hippocampus in Alzheimer’s disease and amnestic mild cognitive impairment. Front Aging Neurosci. 2021;13: 705030.
Xiong Y, Ye C, Chen Y, Zhong X, Chen H, Sun R, et al. Altered functional connectivity of basal ganglia in mild cognitive impairment and Alzheimer’s disease. Brain Sci. 2022;12(11):1555.
Langlais PJ, Thai L, Hansen L, Galasko D, Alford M, Masliah E. Neurotransmitters in basal ganglia and cortex of Alzheimer’s disease with and without Lewy bodies. Neurology. 1993;43(10):1927.
Zhou B, Liu Y, Zhang Z, An N, Yao H, Wang P, et al. Impaired functional connectivity of the thalamus in Alzheimer’s disease and mild cognitive impairment: a resting-state fMRI study. Curr Alzheimer Res. 2013;10(7):754–66.
Moretti D, Paternicò D, Binetti G, Zanetti O, Frisoni GB. EEG markers are associated to gray matter changes in thalamus and basal ganglia in subjects with mild cognitive impairment. Neuroimage. 2012;60(1):489–96.
Liu J, Zhang X, Yu C, Duan Y, Zhuo J, Cui Y, et al. Impaired parahippocampus connectivity in mild cognitive impairment and Alzheimer’s disease. J Alzheimer’s Dis. 2016;49(4):1051–64.
Echávarri C, Aalten P, Uylings HB, Jacobs H, Visser PJ, Gronenschild E, et al. Atrophy in the parahippocampal gyrus as an early biomarker of Alzheimer’s disease. Brain Struct Funct. 2011;215:265–71.
Misra C, Fan Y, Davatzikos C. Baseline and longitudinal patterns of brain atrophy in MCI patients, and their use in prediction of short-term conversion to AD: results from ADNI. Neuroimage. 2009;44(4):1415–22.
Joko T, Washizuka S, Sasayama D, Inuzuka S, Ogihara T, Yasaki T, et al. Patterns of hippocampal atrophy differ among Alzheimer’s disease, amnestic mild cognitive impairment, and late-life depression. Psychogeriatrics. 2016;16(6):355–61.
Barnes J, Scahill RI, Schott JM, Frost C, Rossor MN, Fox NC. Does Alzheimer’s disease affect hippocampal asymmetry? Evidence from a cross-sectional and longitudinal volumetric MRI study. Dementia Geriatr Cogn Disord. 2005;19(5–6):338–44.
Shi F, Liu B, Zhou Y, Yu C, Jiang T. Hippocampal volume and asymmetry in mild cognitive impairment and Alzheimer’s disease: meta-analyses of MRI studies. Hippocampus. 2009;19(11):1055–64.
Yao D, Sui J, Wang M, Yang E, Jiaerken Y, Luo N, et al. A mutual multi-scale triplet graph convolutional network for classification of brain disorders using functional or structural connectivity. IEEE Trans Med Imaging. 2021;40(4):1279–89.
Acknowledgements
This study utilized data from the Alzheimer’s Disease Neuroimaging Initiative (ADNI, available at http://adni.loni.usc.edu) and the Australian Imaging Biomarkers and Lifestyle study (AIBL, available at http://aibl.csiro.au). While the investigators of ADNI and AIBL played a role in designing and implementing these studies and provided the data, they did not directly contribute to the work presented in this manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (61802330, 61802331), Natural Science Foundation of Shandong Province (ZR2020QH048) and Yantai City Science and Technology Innovation Development Plan (2023XDRH006).
Author information
Authors and Affiliations
Contributions
Chuanzhen Zhu and Honglun Li: Conceptualization, Methodology, Data curation, Software, Experiments, and Writing—Original Draft. Zhiwei Song: Methodology, Data curation, Writing—Original Draft. Minbo Jiang: Validation, Formal analysis. Limei Song, Lin Li and Xuan Wang: Writing—Original Draft, Investigation. Qiang Zheng: Conceptualization, Supervision, Project administration and Funding acquisition.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no potential conflicts of interest to disclose.
Ethical approval
This study has ethical approval, consent to participate, and consent for publication. The Institutional Review Board of all participating centers approved the study procedures.
Informed consent
This study has consent to participate.
Consent for publication
This study has consent for publication.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhu, C., Li, H., Song, Z. et al. Jointly constrained group sparse connectivity representation improves early diagnosis of Alzheimer’s disease on routinely acquired T1-weighted imaging-based brain network. Health Inf Sci Syst 12, 19 (2024). https://doi.org/10.1007/s13755-023-00269-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13755-023-00269-0