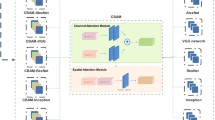
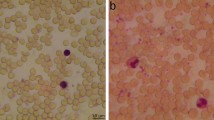
Abstract
This paper considers a new method for providing a recommendation (second opinion) for a laboratory assistant in manual blood typing based on serological plates. The manual method consists of two steps: preparation and analysis. During preparation step the laboratory assistant needs to fill each well of a plate with a blood sample and a reagent mixture according to methodological guidelines. In the second step it is necessary to visually determine the result of the reactions, named agglutination. Despite the popularity of this method, it is slow and highly influenced by human factor, which cause blood typing errors. To increase the quality and performance of the analysis step, we propose a novel neural-based classification method. Our solution provides a fast way to fill the results into a laboratory system. We collected a new large dataset consisting of 3139 well images with GTs from donors’ medical history and six experts’ assessment for each. We showed that the proposed solution based on state-of-the-art architectures is comparable with the best expert and has 2.75 times fewer errors than the average one, with an overall accuracy equal to 98.4%. Taking into account the low-semantic nature of the task, we also considered shallow neural networks, which showed accuracy comparable with state-of-the-art models.







Similar content being viewed by others
Data availability
A test portion of the dataset and sample code will be publicly available once the journal accepts publication.
Notes
Antigens within one system share structural and genetic similarities.
References
International society of blood transfusion (isbt) (2023) https://www.isbtweb.org/
Lee E, Schiffer C. Abo compatibility can influence the results of platelet transfusion: results of a randomized trial. Transfusion. 1989;29(5):384–9. https://doi.org/10.1046/j.1537-2995.1989.29589284135.x.
Ferraz A. 2013 IEEE 3rd Portuguese meeting in bioengineering (ENBENG) (IEEE). 2013, pp. 1–6. https://doi.org/10.1109/enbeng.2013.6518441
Rahman S, Rahman MA, Khan FA, Shahjahan SB, Nahar K. Blood group detection using image processing techniques. Ph.D. thesis, BRAC University, 2017
Sahastrabuddhe AP, Ajij S. Blood group detection and RBC, WBC counting: an image processing approach. Int J Eng Comput Sci. 2016;5(10):18635–9. https://doi.org/10.18535/IJECS/V5I10.49.
Yamin A, Imran F, Akbar U, Tanvir SH. 2017 International conference on communication, computing and digital systems (C-CODE) (IEEE), 2017, pp. 293–298. https://doi.org/10.1109/c-code.2017.7918945
Atici H, Koçer HE, Kader S. Determination of blood group by image processing using digital images. Bilecik Şeyh Edebali Üniversitesi Fen Bilimleri Dergisi. 2020;7(2):649–59. https://doi.org/10.35193/bseufbd.646847.
Dannana S, Prasad DYV. Blood group detection using ml classifier. Int J Health Sci. 2022;6(S1):4395–408. https://doi.org/10.53730/ijhs.v6nS1.5830.
Wang B, Wang L, Xu W, Ding S, Duan S, Cheng W. International Conference on Service Science. New York: Springer; 2023. pp. 231–246.
Odeh N, Toma A, Mohammed F, Dama Y, Oshaibi F, Shaar M. An efficient system for automatic blood type determination based on image matching techniques. Appl Sci. 2021;11(11):5225–51. https://doi.org/10.3390/app11115225.
Sklavounos AA, Lamanna J, Modi D, Gupta S, Mariakakis A, Callum J, Wheeler AR. Digital microfluidic hemagglutination assays for blood typing, donor compatibility testing, and hematocrit analysis. Clin Chem. 2021;67(12):1699–708.
Gupta S, Sklavounos AA, Dahmer J, Yong AK, Abdullah MA, Camacho G, Morton K, Shiu M, Labrecque J, Veres T, et al. 2022 IEEE International Conference on Pervasive Computing and Communications Workshops and other Affiliated Events (PerCom Workshops) (IEEE); 2022. pp. 230–235.
Korchagin SA, Zaychenkova EE, Sharapov DA, Ershov EI, Butorin YV, Vengerov YY. The algorithm of blood typing using serological plates images. Comput Opt. 2023. https://doi.org/10.18287/2412-6179-CO-1339.
Reid ME, Lomas-Francis C, Olsson ML. The blood group antigen factsbook. Cambridge: Academic Press; 2012. https://doi.org/10.1016/C2011-0-69689-9.
Bloody well repository. https://github.com/createcolor/bloody_well (2024)
Tan M, Le Q. International conference on machine learning (PMLR), 2019; 6105–6114. https://doi.org/10.48550/arXiv.1905.11946
Tan M, Le Q. International conference on machine learning. Proc Mach Learn Res PMLR. 2021. https://doi.org/10.48550/arXiv.2104.00298.
Howard A, Sandler M, Chu G, Chen LC, Chen B, Tan M, Wang W, Zhu Y, Pang R, Vasudevan V, et al. Proceedings of the IEEE/CVF international conference on computer vision. 2019, pp. 1314–1324. https://doi.org/10.1109/iccv.2019.00140
Huang G, Liu Z, Van Der Maaten L, Weinberger KQ. Proceedings of the IEEE conference on computer vision and pattern recognition. 2017. pp. 4700–4708. https://doi.org/10.1109/cvpr.2017.243
He K, Zhang X, Ren S, Sun J. Proceedings of the IEEE conference on computer vision and pattern recognition. 2016. pp. 770–778. https://doi.org/10.1109/CVPR.2016.90
Shankar K, Zhang Y, Liu Y, Wu L, Chen CH. Hyperparameter tuning deep learning for diabetic retinopathy fundus image classification. IEEE Access. 2020;8:118164–73. https://doi.org/10.1109/access.2020.3005152.
Pacheco AGC, Krohling RA. An attention-based mechanism to combine images and metadata in deep learning models applied to skin cancer classification. IEEE J Biomed Health Inform. 2021;25(9):3554–63. https://doi.org/10.1109/JBHI.2021.3062002.
Liu D, Wang W, Wu X, Yang J. 2022 3rd International Conference on Electronic Communication and Artificial Intelligence (IWECAI) (IEEE). 2022. pp. 384–387. https://doi.org/10.1109/IWECAI55315.2022.00081
Kingma DP, Ba J. Adam: a method for stochastic optimization. 2014. arXiv:1412.6980. https://doi.org/10.48550/arXiv.1412.6980
Simonyan K, Zisserman A. Very deep convolutional networks for large-scale image recognition. 2014. arXiv:1409.1556. https://doi.org/10.48550/arXiv.1409.1556
Acknowledgements
The authors are sincerely grateful to Yury Butorin for consultations on issues in serology field and help with the dataset collection.
Funding
The study was done with a support of the state assignment of IO RAS (theme No. FFNU-2024- 0025).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have no Conflict of interest to declare that are relevant to the content of this article.
Ethical approval
The study was carried out in accordance with the Declaration of Helsinki (2013). The experimental procedure was approved by the ethics board of the Institute for Information Transmission Problems. The data is completely anonymized and can be published.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Korchagin, S., Zaychenkova, E., Ershov, E. et al. Image-based second opinion for blood typing. Health Inf Sci Syst 12, 28 (2024). https://doi.org/10.1007/s13755-024-00289-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13755-024-00289-4