Abstract
Purpose
Target-based strategy is a prevalent means of drug research and development (R&D), since targets provide effector molecules of drug action and offer the foundation of pharmacological investigation. Recently, the artificial intelligence (AI) technology has been utilized in various stages of drug R&D, where AI-assisted experimental methods show higher efficiency than sole experimental ones. It is a critical need to give a comprehensive review of AI applications in drug R &D for biopharmaceutical field.
Methods
Relevant literatures about AI-assisted drug R&D were collected from the public databases (Including Google Scholar, Web of Science, PubMed, IEEE Xplore Digital Library, Springer, and ScienceDirect) through a keyword searching strategy with the following terms [(“Artificial Intelligence” OR “Knowledge Graph” OR “Machine Learning”) AND (“Drug Target Identification” OR “New Drug Development”)].
Results
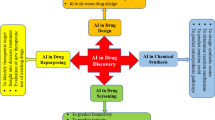
In this review, we first introduced common strategies and novel trends of drug R&D, followed by characteristic description of AI algorithms widely used in drug R&D. Subsequently, we depicted detailed applications of AI algorithms in target identification, lead compound identification and optimization, drug repurposing, and drug analytical platform construction. Finally, we discussed the challenges and prospects of AI-assisted methods for drug discovery.
Conclusion
Collectively, this review provides comprehensive overview of AI applications in drug R&D and presents future perspectives for biopharmaceutical field, which may promote the development of drug industry.






Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.References
Duarte Y, Márquez-Miranda V, Miossec MJ, González-Nilo F. Integration of target discovery, drug discovery and drug delivery: a review on computational strategies. Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2019;11(4):e1554. https://doi.org/10.1002/wnan.1554.
Gashaw I, Ellinghaus P, Sommer A, Asadullah K. What makes a good drug target? Drug Discov Today. 2011;16(23–24):1037–43. https://doi.org/10.1016/j.drudis.2011.09.007.
Csermely P, Agoston V, Pongor S. The efficiency of multi-target drugs: the network approach might help drug design. Trends Pharmacol Sci. 2005;26(4):178–82. https://doi.org/10.1016/j.tips.2005.02.007.
Fan K, Cheng L, Li L. Artificial intelligence and machine learning methods in predicting anti-cancer drug combination effects. Briefings Bioinf. 2021;22(6):bbab271. https://doi.org/10.1093/bib/bbab271.
Mak K-K, Pichika MR. Artificial intelligence in drug development: present status and future prospects. Drug Discov Today. 2019;24(3):773–80. https://doi.org/10.1016/j.drudis.2018.11.014.
Li G, Lin P, Wang K, Gu C-C, Kusari S. Artificial intelligence-guided discovery of anticancer lead compounds from plants and associated microorganisms. Trends Cancer. 2022;8(1):65–80. https://doi.org/10.1016/j.trecan.2021.10.002.
Tolios A, De Las Rivas J, Hovig E, Trouillas P, Scorilas A, Mohr T. Computational approaches in cancer multidrug resistance research: identification of potential biomarkers, drug targets and drug-target interactions. Drug Res Updat: Rev Comment Antimicrob Anticancer Chemother. 2020;48:100662. https://doi.org/10.1016/j.drup.2019.100662.
Zhou Y, Zhang Y, Lian X, Li F, Wang C, Zhu F, Qiu Y, Chen Y. Therapeutic target database update 2022: facilitating drug discovery with enriched comparative data of targeted agents. Nucleic Acids Res. 2022;50(D1):D1398–407. https://doi.org/10.1093/nar/gkab953.
Wang L, Ding J, Pan L, Cao D, Jiang H, Ding X. Artificial intelligence facilitates drug design in the big data era. Chemom Intell Lab Syst. 2019;194:103850. https://doi.org/10.1016/j.chemolab.2019.103850.
Liu G, Xie Y, Sun Y, Zhang K, Ma J, Huang Y. Drug research and development opportunities in low- and middle-income countries: accelerating traditional medicine through systematic utilization and comprehensive synergy. Infect Dis Poverty. 2022;11(1):27. https://doi.org/10.1186/s40249-022-00954-4.
Paul D, Sanap G, Shenoy S, Kalyane D, Kalia K, Tekade RK. Artificial intelligence in drug discovery and development. Drug Discov Today. 2021;26(1):80–93. https://doi.org/10.1016/j.drudis.2020.10.010.
Sun D, Gao W, Hu H, Zhou S. Why 90% of clinical drug development fails and how to improve it? Acta Pharmaceutica Sinica B. 2022;12(7):3049–62. https://doi.org/10.1016/j.apsb.2022.02.002.
Takebe T, Imai R, Ono S. The current status of drug discovery and development as originated in united states academia: the influence of industrial and academic collaboration on drug discovery and development. Clin Transl Sci. 2018;11(6):597–606. https://doi.org/10.1111/cts.12577.
Cook D, Brown D, Alexander R, March R, Morgan P, Satterthwaite G, Pangalos MN. Lessons learned from the fate of AstraZeneca’s drug pipeline: a five-dimensional framework. Nat Rev Drug Discov. 2014;13(6):419–31. https://doi.org/10.1038/nrd4309.
Meyers RM, Bryan JG, McFarland JM, Weir BA, Sizemore AE, Xu H, Dharia NV, Montgomery PG, Cowley GS, Pantel S, Goodale A, Lee Y, Ali LD, Jiang G, Lubonja R, Harrington WF, Strickland M, Wu T, Hawes DC, Zhivich VA, Wyatt MR, Kalani Z, Chang JJ, Okamoto M, Stegmaier K, Golub TR, Boehm JS, Vazquez F, Root DE, Hahn WC, Tsherniak A. Computational correction of copy number effect improves specificity of CRISPR-Cas9 essentiality screens in cancer cells. Nat Genet. 2017;49(12):1779–84. https://doi.org/10.1038/ng.3984.
Pinu FR, Beale DJ, Paten AM, Kouremenos K, Swarup S, Schirra HJ, Wishart D. Systems biology and multi-omics integration: viewpoints from the metabolomics research community. Metabolites. 2019;9(4):76. https://doi.org/10.3390/metabo9040076.
Galluzzi L, Vitale I, Michels J, Brenner C, Szabadkai G, Harel-Bellan A, Castedo M, Kroemer G. Systems biology of cisplatin resistance: past, present and future. Cell Death Dis. 2014;5(5):e1257–e1257. https://doi.org/10.1038/cddis.2013.428.
Keseler IM, Mackie A, Peralta-Gil M, Santos-Zavaleta A, Gama-Castro S, Bonavides-Martínez C, Fulcher C, Huerta AM, Kothari A, Krummenacker M, Latendresse M, Muñiz-Rascado L, Ong Q, Paley S, Schröder I, Shearer AG, Subhraveti P, Travers M, Weerasinghe D, Weiss V, Collado-Vides J, Gunsalus RP, Paulsen I, Karp PD. EcoCyc: fusing model organism databases with systems biology. Nucleic Acids Res. 2013;41(D1):D605–12. https://doi.org/10.1093/nar/gks1027.
Gogleva A, Polychronopoulos D, Pfeifer M, Poroshin V, Ughetto M, Martin MJ, Thorpe H, Bornot A, Smith PD, Sidders B, Dry JR, Ahdesmäki M, McDermott U, Papa E, Bulusu KC. Knowledge graph-based recommendation framework identifies drivers of resistance in EGFR mutant non-small cell lung cancer. Nat Commun. 2022;13(1):1667. https://doi.org/10.1038/s41467-022-29292-7.
Chandak P, Huang K, Zitnik M. Building a knowledge graph to enable precision medicine. Sci Data. 2023;10(1):1–16. https://doi.org/10.1038/s41597-023-01960-3.
Zador A, Escola S, Richards B, Ölveczky B, Bengio Y, Boahen K, Botvinick M, Chklovskii D, Churchland A, Clopath C, DiCarlo J, Ganguli S, Hawkins J, Körding K, Koulakov A, LeCun Y, Lillicrap T, Marblestone A, Olshausen B, Pouget A, Savin C, Sejnowski T, Simoncelli E, Solla S, Sussillo D, Tolias AS, Tsao D. Catalyzing next-generation artificial intelligence through NeuroAI. Nat Commun. 2023;14(1):1597.
Gupta RR. Application of artificial intelligence and machine learning in drug discovery. In: Heifetz A, editor. Artificial intelligence in drug design. New York: Springer; 2022. p. 113–24.
Sarkar C, Das B, Rawat VS, Wahlang JB, Nongpiur A, Tiewsoh I, Lyngdoh NM, Das D, Bidarolli M, Sony HT. Artificial intelligence and machine learning technology driven modern drug discovery and development. Int J Mol Sci. 2023;24(3):2026. https://doi.org/10.3390/ijms24032026.
Rashid MBMA. Artificial intelligence effecting a paradigm shift in drug development. SLAS Technol. 2021;26(1):3–15. https://doi.org/10.1177/2472630320956931.
Recanatini M, Cabrelle C. Drug research meets network science: where are we? J Med Chem. 2020;63(16):8653–66. https://doi.org/10.1021/acs.jmedchem.9b01989.
Li D, Hu J, Zhang L, Li L, Yin Q, Shi J, Guo H, Zhang Y, Zhuang P. Deep learning and machine intelligence: new computational modeling techniques for discovery of the combination rules and pharmacodynamic characteristics of traditional Chinese medicine. Eur J Pharmacol. 2022;933:175260. https://doi.org/10.1016/j.ejphar.2022.175260.
She S, Chen H, Ji W, Sun M, Cheng J, Rui M, Feng C. Deep learning-based multi-drug synergy prediction model for individually tailored anti-cancer therapies. Front Pharmacol. 2022;13:1032875. https://doi.org/10.3389/fphar.2022.1032875.
Adams SA, Petersen C. Precision medicine: opportunities, possibilities, and challenges for patients and providers. J Am Med Inf Assoc. 2016;23(4):787–90. https://doi.org/10.1093/jamia/ocv215.
Fukushima K. Neocognitron: a self-organizing neural network model for a mechanism of pattern recognition unaffected by shift in position. Biol Cybern. 1980;36(4):193–202. https://doi.org/10.1007/BF00344251.
Lipton Z. A critical review of recurrent neural networks for sequence learning. 2015.
Hinton GE, Osindero S, Teh Y-W. A fast learning algorithm for deep belief nets. Neural Comput. 2006;18(7):1527–54. https://doi.org/10.1162/neco.2006.18.7.1527.
Gerstein M. ENCODE leads the way on big data. Nature. 2012;489(7415):208–208. https://doi.org/10.1038/489208b.
Goodfellow IJ, Pouget-Abadie J, Mirza M, Xu B, Warde-Farley D, Ozair S, Courville A, Bengio Y. Generative adversarial nets. In: Proceedings of the 27th International Conference on Neural Information Processing Systems—Volume 2, MIT Press: Montreal, Canada, 2014; pp 2672–2680.
Abadi M, Barham P, Chen J, Chen Z, Davis A, Dean J., Devin M, Ghemawat S, Irving G, Isard M, Kudlur M, Levenberg J, Monga R, Moore S, Murray DG, Steiner B, Tucker PA, Vasudevan V, Warden P, Wicke M, Yu Y, Zhang X. In: TensorFlow: a system for large-scale machine learning, USENIX Symposium on Operating Systems Design and Implementation. 2016.
Ii B. The power of deep learning to ligand-based novel drug discovery. Expert Opin Drug Discov. 2020. https://doi.org/10.1080/17460441.2020.1745183.
Bonner S, Barrett IP, Ye C, Swiers R, Engkvist O, Hoyt CT, Hamilton WL. Understanding the performance of knowledge graph embeddings in drug discovery. Artif Intell Life Sci. 2022;2:100036. https://doi.org/10.1016/j.ailsci.2022.100036.
Acosta JN, Falcone GJ, Rajpurkar P, Topol EJ. Multimodal biomedical AI. Nat Med. 2022;28(9):1773–84. https://doi.org/10.1038/s41591-022-01981-2.
Kırboğa KK, Abbasi S, Küçüksille EU. Explainability and white box in drug discovery. Chem Biol Drug Des. 2023;102(1):217–33. https://doi.org/10.1111/cbdd.14262.
Zongsheng W, Xue R, Shao M. Knowledge graph analysis and visualization of AI technology applied in COVID-19. Environ Sci Pollut Res Int. 2022. https://doi.org/10.1007/s11356-021-17800-z.
Galindez G, Matschinske J, Rose TD, Sadegh S, Salgado-Albarrán M, Späth J, Baumbach J, Pauling JK. Lessons from the COVID-19 pandemic for advancing computational drug repurposing strategies. Nat Comput Sci. 2021;1(1):33–41. https://doi.org/10.1038/s43588-020-00007-6.
Maghsoudi S, Taghavi Shahraki B, Rameh F, Nazarabi M, Fatahi Y, Akhavan O, Rabiee M, Mostafavi E, Lima EC, Saeb MR, Rabiee N. A review on computer-aided chemogenomics and drug repositioning for rational COVID-19 drug discovery. Chem Biol Drug Des. 2022;100(5):699–721. https://doi.org/10.1111/cbdd.14136.
Jiang C, Ngo V, Chapman R, Yue Y, Liu H, Jiang G, Zong N. Deep denoising of raw biomedical knowledge graph from COVID-19 literature, LitCovid, and pubtator: framework development and validation. J Med Internet Res. 2022. https://doi.org/10.2196/38584.
Jin W, Stokes JM, Eastman RT, Itkin Z, Zakharov AV, Collins JJ, Jaakkola TS, Barzilay R. Deep learning identifies synergistic drug combinations for treating COVID-19. Proc Natl Acad Sci USA. 2021;118(39):e2105070118. https://doi.org/10.1073/pnas.2105070118.
Verma R, Raj S, Berry U, Ranjith-Kumar CT, Surjit M. Drug repurposing for COVID-19 therapy: pipeline, current status and challenges. In: Sobti RC, Lal SK, Goyal RK, editors. Drug repurposing for emerging infectious diseases and cancer. Singapore: Springer; 2023. p. 451–78.
Ozdemir ES, Ranganathan SV, Nussinov R. How has artificial intelligence impacted COVID-19 drug repurposing and what lessons have we learned? Expert Opin Drug Discov. 2022;17(10):1061–5. https://doi.org/10.1080/17460441.2022.2128333.
Shen WX, Liu Y, Chen Y, Zeng X, Tan Y, Jiang YY, Chen YuZ. AggMapNet: enhanced and explainable low-sample omics deep learning with feature-aggregated multi-channel networks. Nucleic Acids Res. 2022;50(8):e45. https://doi.org/10.1093/nar/gkac010.
Pham T-H, Qiu Y, Zeng J, Xie L, Zhang P. A deep learning framework for high-throughput mechanism-driven phenotype compound screening and its application to COVID-19 drug repurposing. Nat Mach Intell. 2021;3(3):247–57. https://doi.org/10.1038/s42256-020-00285-9.
Ju J, Wismans LV, Mustafa DAM, Reinders MJT, van Eijck CHJ, Stubbs AP, Li Y. Robust deep learning model for prognostic stratification of pancreatic ductal adenocarcinoma patients. iScience. 2021;24(12):103415. https://doi.org/10.1016/j.isci.2021.103415.
Long NP, Jung KH, Anh NH, Yan HH, Nghi TD, Park S, Yoon SJ, Min JE, Kim HM, Lim JH, Kim JM, Lim J, Lee S, Hong S-S, Kwon SW. An integrative data mining and omics-based translational model for the identification and validation of oncogenic biomarkers of pancreatic cancer. Cancers. 2019;11(2):155. https://doi.org/10.3390/cancers11020155.
Wei Q, Ramsey SA. Predicting chemotherapy response using a variational autoencoder approach. BMC Bioinf. 2021;22(1):453. https://doi.org/10.1186/s12859-021-04339-6.
Shuangshuang L, Lin Q, Yun T, Fenghui L. In: A deep learning fusion clustering framework for breast cancer subtypes identification by integrating multi-omics data, 2020 5th International Conference on Mechanical, Control and Computer Engineering (ICMCCE), 2020–12; 2020; pp 1710–1714.
Chai H, Zhou X, Zhang Z, Rao J, Zhao H, Yang Y. Integrating multi-omics data through deep learning for accurate cancer prognosis prediction. Comput Biol Med. 2021;134:104481. https://doi.org/10.1016/j.compbiomed.2021.104481.
Khan D, Shedole S. Leveraging deep learning techniques and integrated omics data for tailored treatment of breast cancer. J Personal Med. 2022;12(5):674. https://doi.org/10.3390/jpm12050674.
Song H, Ruan C, Xu Y, Xu T, Fan R, Jiang T, Cao M, Song J. Survival stratification for colorectal cancer via multi-omics integration using an autoencoder-based model. Exp Biol Med. 2022;247(11):898–909. https://doi.org/10.1177/15353702211065010.
Cao R, Yang F, Ma S-C, Liu L, Zhao Y, Li Y, Wu D-H, Wang T, Lu W-J, Cai W-J, Zhu H-B, Guo X-J, Lu Y-W, Kuang J-J, Huan W-J, Tang W-M, Huang K, Huang J, Yao J, Dong Z-Y. Development and interpretation of a pathomics-based model for the prediction of microsatellite instability in colorectal cancer. Theranostics. 2020;10(24):11080–91. https://doi.org/10.7150/thno.49864.
Rong Z, Liu Z, Song J, Cao L, Yu Y, Qiu M, Hou Y. MCluster-VAEs: an end-to-end variational deep learning-based clustering method for subtype discovery using multi-omics data. Comput Biol Med. 2022;150:106085. https://doi.org/10.1016/j.compbiomed.2022.106085.
Schulte-Sasse R, Budach S, Hnisz D, Marsico A. Integration of multiomics data with graph convolutional networks to identify new cancer genes and their associated molecular mechanisms. Nat Mach Intell. 2021;3(6):513–26. https://doi.org/10.1038/s42256-021-00325-y.
Wang Y, Yang Y, Chen S, Wang J. DeepDRK: a deep learning framework for drug repurposing through kernel-based multi-omics integration. Briefings Bioinf. 2021;22(5):bbab048. https://doi.org/10.1093/bib/bbab048.
Strogatz SH. Exploring complex networks. Nature. 2001;410(6825):268–76. https://doi.org/10.1038/35065725.
Barabasi AL, Oltvai ZN. Network biology: understanding the cell’s functional organization. Nat Rev Genet. 2004;5(2):101–13. https://doi.org/10.1038/nrg1272.
Have CT, Jensen LJ. Are graph databases ready for bioinformatics? Bioinformatics. 2013;29(24):3107–8. https://doi.org/10.1093/bioinformatics/btt549.
Balaur I, Mazein A, Saqi M, Lysenko A, Rawlings CJ, Auffray C. Recon2Neo4j: applying graph database technologies for managing comprehensive genome-scale networks. Bioinformatics. 2017;33(7):1096–8. https://doi.org/10.1093/bioinformatics/btw731.
Yan J, Risacher SL, Shen L, Saykin AJ. Network approaches to systems biology analysis of complex disease: integrative methods for multi-omics data. Brief Bioinform. 2018;19(6):1370–81. https://doi.org/10.1093/bib/bbx066.
Barabasi AL, Gulbahce N, Loscalzo J. Network medicine: a network-based approach to human disease. Nat Rev Genet. 2011;12(1):56–68. https://doi.org/10.1038/nrg2918.
Hogan A, Blomqvist E, Cochez M, d’Amato C, de Melo G, Gutierrez C, Gayo JEL, Kirrane S, Neumaier S, Polleres A, Navigli R, Ngomo A-CN, Rashid SM, Rula A, Schmelzeisen L, Sequeda J, Staab S, Zimmermann A. Knowledge graphs. ACM Comput Surv. 2022;54(4):1–37. https://doi.org/10.1145/3447772.
CFP: special issue on knowledge graphs.
Walsh B, Mohamed SK, Nováček V. In: BioKG: a knowledge graph for relational learning on biological data, association for computing machinery. 2020 pp 3173–3180.
Shin J, Piao Y, Bang D, Kim S, Jo K. DRPreter: interpretable anticancer drug response prediction using knowledge-guided graph neural networks and transformer. Int J Mol Sci. 2022. https://doi.org/10.3390/ijms232213919.
Himmelstein DS, Lizee A, Hessler C, Brueggeman L, Chen SL, Hadley D, Green A, Khankhanian P, Baranzini SE. Systematic integration of biomedical knowledge prioritizes drugs for repurposing. Elife. 2017;6:e26726. https://doi.org/10.7554/eLife.26726.
Liu Z, Chen Q, Lan W, Liang J, Chen YP, Chen B. A survey of network embedding for drug analysis and prediction. Curr Protein Pept Sci. 2020. https://doi.org/10.2174/1389203721666200702145701.
Zhang R, Hristovski D, Schutte D, Kastrin A, Fiszman M, Kilicoglu H. Drug repurposing for COVID-19 via knowledge graph completion. J Biomed Inf. 2021;115:103696. https://doi.org/10.1016/j.jbi.2021.103696.
Sun Z, Deng ZH, Nie JY, Tang J. RotatE: knowledge graph embedding by relational rotation in complex space. [Preprint] 2019 Available from: https://doi.org/10.48550/arXiv.1902.10197
Novak TP, Hoffman DL. Residual scaling: an alternative to correspondence analysis for the graphical representation of residuals from log-linear models. Multivar Behav Res. 1990;25(3):351–70. https://doi.org/10.1207/s15327906mbr2503_7.
Al-Obeidat F, Rocha Á, Khan MS, Maqbool F, Razzaq S. Parallel tensor factorization for relational learning. Neural Comput Appl. 2022;34(11):8455–64. https://doi.org/10.1007/s00521-021-05692-6.
Chachlakis DG, Tsitsikas Y, Papalexakis EE, Markopoulos PP. In: Robust multi-relational learning with absolute projection rescal, 2019 IEEE Global Conference on Signal and Information Processing (GlobalSIP), 2019–11; 2019; pp 1–5.
Cai B, Xiang Y, Gao L, Wu D, Zhang H, Jin J, Luan T. From wide to deep: dimension lifting network for parameter-efficient knowledge graph embedding. [Preprint] 2023 Available from: https://doi.org/10.48550/arXiv.2303.12816
Xie Z, Zhu R, Zhang M, Liu J. In: SparseMult: a tensor decomposition model based on sparse relation matrix, 2022 IEEE/WIC/ACM International Joint Conference on Web Intelligence and Intelligent Agent Technology (WI-IAT), 2022–11; 2022; pp 761–764.
Ramos CCO, Rodrigues D, de Souza AN, Papa JP. On the study of commercial losses in Brazil: a binary black hole algorithm for theft characterization. IEEE Trans Smart Grid. 2018;9(2):676–83. https://doi.org/10.1109/TSG.2016.2560801.
Kacha L, Zitouni A, Djoudi M. KAB: a new k-anonymity approach based on black hole algorithm. J King Saud Univ: Comput Inform Sci. 2022;34(7):4075–88. https://doi.org/10.1016/j.jksuci.2021.04.014.
Hatamlou A. Black hole: a new heuristic optimization approach for data clustering. Inf Sci. 2013;222:175–84. https://doi.org/10.1016/j.ins.2012.08.023.
Yao L, Mao C, Luo Y. KG-BERT: BERT for knowledge graph completion. [Preprint] 2019 Available from: https://doi.org/10.48550/arXiv.1909.03193
Cabalar P, Diéguez M. STeLP—a tool for temporal answer set programming. In: Delgrande JP, Faber W, editors. Logic programming and nonmonotonic reasoning. Cham: Springer; 2011. p. 370–5.
Patel L, Shukla T, Huang X, Ussery DW, Wang S. Machine learning methods in drug discovery. Molecules. 2020. https://doi.org/10.3390/molecules25225277.
Cheng F, Zhao Z. Machine learning-based prediction of drug-drug interactions by integrating drug phenotypic, therapeutic, chemical, and genomic properties. J Am Med Inform Assoc. 2014;21(e2):e278-286. https://doi.org/10.1136/amiajnl-2013-002512.
Usha T, Shanmugarajan D, Goyal AK, Kumar CS, Middha SK. Recent updates on computer-aided drug discovery: time for a paradigm shift. Curr Top Med Chem. 2017;17(30):3296–307. https://doi.org/10.2174/1568026618666180101163651.
Maia EHB, Assis LC, de Oliveira TA, da Silva AM, Taranto AG. Structure-based virtual screening: from classical to artificial intelligence. Front Chem. 2020. https://doi.org/10.3389/fchem.2020.00343.
Staszak M, Staszak K, Wieszczycka K, Bajek A, Roszkowski K, Tylkowski B. Machine learning in drug design: use of artificial intelligence to explore the chemical structure-biological activity relationship. Wiley Interdiscip Rev-Comput Mol Sci. 2022. https://doi.org/10.1002/wcms.1568.
Zulkifli MH, Abdullah ZL, Mohamed Yusof NIS, Mohd Fauzi F. In silico toxicity studies of traditional Chinese herbal medicine: a mini review. Curr Opin Struct Biol. 2023;80:102588.
Canzler S, Schor J, Busch W, Schubert K, Rolle-Kampczyk UE, Seitz H, Kamp H, von Bergen M, Buesen R, Hackermüller J. Prospects and challenges of multi-omics data integration in toxicology. Arch Toxicol. 2020;94(2):371–88. https://doi.org/10.1007/s00204-020-02656-y.
Wu Q, Cai C, Guo P, Chen M, Wu X, Zhou J, Luo Y, Zou Y, Liu A-L, Wang Q, Kuang Z, Fang J. In silico identification and mechanism exploration of hepatotoxic ingredients in traditional Chinese medicine. Front Pharmacol. 2019;10:458. https://doi.org/10.3389/fphar.2019.00458.
Conn JGM, Carter JW, Conn JJA, Subramanian V, Baxter A, Engkvist O, Llinas A, Ratkova EL, Pickett SD, McDonagh JL, Palmer DS. Blinded predictions and post hoc analysis of the second solubility challenge data: exploring training data and feature set selection for machine and deep learning models. J Chem Inf Model. 2023;63(4):1099–113. https://doi.org/10.1021/acs.jcim.2c01189.
Cysewski P, Jeliński T, Przybyłek M, Nowak W, Olczak M. Solubility characteristics of acetaminophen and phenacetin in binary mixtures of aqueous organic solvents: experimental and deep machine learning screening of green dissolution media. Pharmaceutics. 2022;14(12):2828. https://doi.org/10.3390/pharmaceutics14122828.
Surov AO, Ramazanova AG, Voronin AP, Drozd KV, Churakov AV, Perlovich GL. Virtual screening, structural analysis, and formation thermodynamics of carbamazepine cocrystals. Pharmaceutics. 2023;15(3):836.
Khan AKA, Malim NHAH. Comparative studies on resampling techniques in machine learning and deep learning models for drug-target interaction prediction. Molecules (Basel, Switzerland). 2023;28(4):1663. https://doi.org/10.3390/molecules28041663.
Ma W, Zhang S, Li Z, Jiang M, Wang S, Guo N, Li Y, Bi X, Jiang H, Wei Z. Predicting drug-target affinity by learning protein knowledge from biological networks. IEEE J Biomed Health Inf. 2023;27(4):2128–37. https://doi.org/10.1109/JBHI.2023.3240305.
Bian J, Zhang X, Zhang X, Xu D, Wang G. MCANet: shared-weight-based MultiheadCrossAttention network for drug-target interaction prediction. Briefings Bioinf. 2023;24(2):bbad082. https://doi.org/10.1093/bib/bbad082.
Firoz A, Malik A, Ali HM, Akhter Y, Manavalan B, Kim C-B. PRR-HyPred: A two-layer hybrid framework to predict pattern recognition receptors and their families by employing sequence encoded optimal features. Int J Biol Macromol. 2023;234:123622. https://doi.org/10.1016/j.ijbiomac.2023.123622.
Bugnon LA, Fenoy E, Edera AA, Raad J, Stegmayer G, Milone DH. Transfer learning: the key to functionally annotate the protein universe. Patterns. 2023;4(2):100691. https://doi.org/10.1016/j.patter.2023.100691.
Sanderson T, Bileschi ML, Belanger D, Colwell LJ. ProteInfer, deep neural networks for protein functional inference. Elife. 2023;12:e80942. https://doi.org/10.7554/eLife.80942.
Hsieh KL, Wang YY, Chen LY, Zhao ZM, Savitz S, Jiang XQ, Tang J, Kim YJ. Drug repurposing for COVID-19 using graph neural network and harmonizing multiple evidence. Sci Rep. 2021. https://doi.org/10.1038/s41598-021-02353-5.
Ren ZH, Yu CQ, Li LP, You ZH, Guan YJ, Wang XF, Pan J. BioDKG-DDI: predicting drug-drug interactions based on drug knowledge graph fusing biochemical information. Brief Funct Genomics. 2022;21(3):216–29. https://doi.org/10.1093/bfgp/elac004.
Zhang J, Chen M, Liu J, Peng DD, Dai Z, Zou XY, Li ZC. A Knowledge-graph-based multimodal deep learning framework for identifying drug-drug interactions. Molecules. 2023. https://doi.org/10.3390/molecules28031490.
Chen S, Semenov I, Zhang F, Yang Y, Geng J, Feng X, Meng Q, Lei K. An effective framework for predicting drug-drug interactions based on molecular substructures and knowledge graph neural network. Comput Biol Med. 2023;169:107900. https://doi.org/10.1016/j.compbiomed.2023.107900.
Lin, X., Quan, Z., Wang, Z.-J., Ma, T., Zeng, X. In KGNN: knowledge graph neural network for drug-drug interaction prediction, (2021). pp 2739–2745. Accesssed 01 July 2021
Zhang S, Lin X, Zhang X. Discovering DTI and DDI by Knowledge Graph with MHRW and Improved Neural Network. In 2021 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), IEEE: 2021.
Zhu J, Sova P, Xu QW, Dombek KM, Xu EY, Vu H, Tu ZD, Brem RB, Bumgarner RE, Schadt EE. Stitching together multiple data dimensions reveals interacting metabolomic and transcriptomic networks that modulate cell regulation. PLoS Biol. 2012. https://doi.org/10.1371/journal.pbio.1001301.
Bartel J, Krumsiek J, Schramm K, Adamski J, Gieger C, Herder C, Carstensen M, Peters A, Rathmann W, Roden M, Strauch K, Suhre K, Kastenmuller G, Prokisch H, Theis FJ. The human blood metabolome-transcriptome interface. PLoS Genet. 2015;11(6):e1005274. https://doi.org/10.1371/journal.pgen.1005274.
Huang JL, Niu CQ, Green CD, Yang L, Mei HK, Han JDJ. Systematic prediction of pharmacodynamic drug-drug interactions through protein-protein-interaction network. PLoS Comput Biol. 2013. https://doi.org/10.1371/journal.pcbi.1002998.
Bonnet E, Calzone L, Michoel T. Integrative multi-omics module network inference with Lemon-Tree. PLoS Comput Biol. 2015. https://doi.org/10.1371/journal.pcbi.1003983.
Mohamed SK, Novácek V, Nounu A. Discovering protein drug targets using knowledge graph embeddings. Bioinformatics. 2020;36(2):603–10. https://doi.org/10.1093/bioinformatics/btz600.
Kim S, Thapa I, Ali H, A multi-omics graph database for data integration and knowledge extraction. In: Proceedings of the 13th ACM International Conference on Bioinformatics, Computational Biology and Health Informatics, ACM: 2022.
Santos A, Colaço AR, Nielsen AB, Niu L, Strauss M, Geyer PE, Coscia F, Albrechtsen NJW, Mundt F, Jensen LJ, Mann M. A knowledge graph to interpret clinical proteomics data. Nat Biotechnol. 2022;40(5):692–702. https://doi.org/10.1038/s41587-021-01145-6.
Zhang N, Bi Z, Liang X, Cheng S, Hong H, Deng S, Lian J, Zhang Q, Chen H, OntoProtein: protein pretraining with gene ontology embedding. arXiv: 2022.
Zhang P, Wang F, Hu JY, Sorrentino R. Label propagation prediction of drug-drug interactions based on clinical side effects. Sci Rep. 2015. https://doi.org/10.1038/srep12339.
Peng W, Chen TL, Liu HC, Dai W, Yu N, Lan W. Improving drug response prediction based on two-space graph convolution. Comput Biol Med. 2023. https://doi.org/10.1016/j.compbiomed.2023.106859.
Boobier S, Hose DRJ, Blacker AJ, Nguyen BN. Machine learning with physicochemical relationships: solubility prediction in organic solvents and water. Nat Commun. 2020;11(1):5753. https://doi.org/10.1038/s41467-020-19594-z.
Nguyen T, Le H, Quinn TP, Nguyen T, Le TD, Venkatesh S. GraphDTA: predicting drug-target binding affinity with graph neural networks. Bioinformatics. 2021;37(8):1140–7. https://doi.org/10.1093/bioinformatics/btaa921.
Bileschi ML, Belanger D, Bryant D, Sanderson T, Carter B, Sculley D, Bateman A, DePristo MA, Colwell LJ. Using deep learning to annotate the protein universe. Nat Biotechnol. 2022;40(6):932. https://doi.org/10.1038/s41587-021-01179-w.
Wang H, Huang F, Xiong Z, Zhang W. A heterogeneous network-based method with attentive mea-path extraction for predicting drug-target interactions. Briefings Bioinf. 2022;23(4):bbac184. https://doi.org/10.1093/bib/bbac184.
Nath A, Kumari P, Chaube R. Prediction of human drug targets and their interactions using machine learning methods: current and future perspectives. Methods Mol Biol. 2018;1762:21–30. https://doi.org/10.1007/978-1-4939-7756-7_2.
Csermely P, Korcsmáros T, Kiss HJM, London G, Nussinov R. Structure and dynamics of molecular networks: a novel paradigm of drug discovery. Pharmacol Ther. 2013;138(3):333–408. https://doi.org/10.1016/j.pharmthera.2013.01.016.
Fathima S, Sinha S, Donakonda S. Network analysis identifies drug targets and small molecules to modulate apoptosis resistant cancers. Cancers. 2021;13(4):851. https://doi.org/10.3390/cancers13040851.
Li X, Ma J, Leng L, Han M, Li M, He F, Zhu Y. MoGCN: a multi-omics integration method based on graph convolutional network for cancer subtype analysis. Front Genet. 2022;13:806842.
Yoon B-H, Kim S-K, Kim S-Y. Use of graph database for the integration of heterogeneous biological data. Genomics Inform. 2017;15(1):19–27. https://doi.org/10.5808/GI.2017.15.1.19.
Paliwal S, de Giorgio A, Neil D, Michel J-B, Lacoste AM. Preclinical validation of therapeutic targets predicted by tensor factorization on heterogeneous graphs. Sci Rep. 2020;10(1):18250. https://doi.org/10.1038/s41598-020-74922-z.
Ji B-Y, You Z-H, Cheng L, Zhou J-R, Alghazzawi D, Li L-P. Predicting miRNA-disease association from heterogeneous information network with GraRep embedding model. Sci Rep. 2020;10(1):6658. https://doi.org/10.1038/s41598-020-63735-9.
Lan W, Wang J, Li M, Liu J, Li Y, Wu F-X, Pan Y. Predicting drug–target interaction using positive-unlabeled learning. Neurocomputing. 2016;206:50–7. https://doi.org/10.1016/j.neucom.2016.03.080.
Nguyen T, Le H, Quinn TP, Nguyen T, Le TD. and Venkatesh, SGraphDTA: predicting drug-target binding affinity with graph neural networks. Bioinformatics. 2021. https://doi.org/10.1093/bioinformatics/btaa921.
Lin X, Li X, Lin X. A Review on applications of computational methods in drug screening and design. Molecules. 2020;25(6):1375. https://doi.org/10.3390/molecules25061375.
Schneider P, Schneider G. De-orphaning the marine natural product (±)-marinopyrrole A by computational target prediction and biochemical validation. Chem Commun. 2017;53(14):2272–4. https://doi.org/10.1039/C6CC09693J.
Cockroft NT, Cheng X, Fuchs JR. STarFish: a stacked ensemble target fishing approach and its application to natural products. J Chem Inf Model. 2019;59(11):4906–20. https://doi.org/10.1021/acs.jcim.9b00489.
Patridge E, Gareiss P, Kinch MS, Hoyer D. An analysis of FDA-approved drugs: natural products and their derivatives. Drug Discov Today. 2016;21(2):204–7. https://doi.org/10.1016/j.drudis.2015.01.009.
Friedrich L, Cingolani G, Ko Y-H, Iaselli M, Miciaccia M, Perrone MG, Neukirch K, Bobinger V, Merk D, Hofstetter RK, Werz O, Koeberle A, Scilimati A, Schneider G. Learning from nature: from a marine natural product to synthetic cyclooxygenase-1 inhibitors by automated de novo design. Adv Sci. 2021;8(16):e2100832. https://doi.org/10.1002/advs.202100832.
Vineetha S, Bhat CCS, Idicula SM. MicroRNA–mRNA interaction network using TSK-type recurrent neural fuzzy network. Gene. 2013;515(2):385–90. https://doi.org/10.1016/j.gene.2012.12.063.
Kwon M-S, Kim Y, Lee S, Namkung J, Yun T, Yi SG, Han S, Kang M, Kim SW, Jang J-Y, Park T. Integrative analysis of multi-omics data for identifying multi-markers for diagnosing pancreatic cancer. BMC Genomics. 2015;16(9):1–10. https://doi.org/10.1186/1471-2164-16-S9-S4.
Madhukar NS, Khade PK, Huang L, Gayvert K, Galletti G, Stogniew M, Allen JE, Giannakakou P, Elemento O. A Bayesian machine learning approach for drug target identification using diverse data types. Nat Commun. 2019;10(1):5221. https://doi.org/10.1038/s41467-019-12928-6.
Arora P, Adams CH, Gudelsky G, DasGupta B, Desai PB. Plasma and brain pharmacokinetics of letrozole and drug interaction studies with temozolomide in NOD-scid gamma mice and sprague dawley rats. Cancer Chemother Pharmacol. 2019;83(1):81–9. https://doi.org/10.1007/s00280-018-3705-6.
Skalic M, Jiménez J, Sabbadin D, De Fabritiis G. Shape-based generative modeling for de novo drug design. J Chem Inf Model. 2019;59(3):1205–14. https://doi.org/10.1021/acs.jcim.8b00706.
Kang S, Cho K. Conditional molecular design with deep generative models. J Chem Inf Model. 2019;59(1):43–52. https://doi.org/10.1021/acs.jcim.8b00263.
Ståhl N, Falkman G, Karlsson A, Mathiason G, Boström J. Deep reinforcement learning for multiparameter optimization in de novo drug design. J Chem Inf Model. 2019;59(7):3166–76. https://doi.org/10.1021/acs.jcim.9b00325.
Liu X, Ye K, van Vlijmen HWT, Izerman AP, van Westen GJP. DrugEx v3: scaffold-constrained drug design with graph transformer-based reinforcement learning. J Cheminf. 2023;15(1):24. https://doi.org/10.1186/s13321-023-00694-z.
Bos PH, Houang EM, Ranalli F, Leffler AE, Boyles NA, Eyrich VA, Luria Y, Katz D, Tang H, Abel R, Bhat S. AutoDesigner, a de novo design algorithm for rapidly exploring large chemical space for lead optimization: application to the design and synthesis of d-amino acid oxidase inhibitors. J Chem Inf Model. 2022;62(8):1905–15. https://doi.org/10.1021/acs.jcim.2c00072.
Langedijk J, Mantel-Teeuwisse AK, Slijkerman DS, Schutjens M-HDB. Drug repositioning and repurposing: terminology and definitions in literature. Drug Discov Today. 2015;20(8):1027–34. https://doi.org/10.1016/j.drudis.2015.05.001.
Nowak-Sliwinska P, Scapozza L, Ruiz i Altaba A. Drug repurposing in oncology compounds, pathways phenotypes and computational approaches for colorectal cancer. Biochim et Biophys Acta (BBA): Rev Cancer. 2019;1871(2):434–54.
Palve V, Liao Y, Remsing Rix LL, Rix U. Turning liabilities into opportunities: off-target based drug repurposing in cancer. Semin Cancer Biol. 2021;68:209–29. https://doi.org/10.1016/j.semcancer.2020.02.003.
Huang Y, Hsu JC, Koo H, Cormode DP. Repurposing ferumoxytol: diagnostic and therapeutic applications of an FDA-approved nanoparticle. Theranostics. 2022;12(2):796–816. https://doi.org/10.7150/thno.67375.
Zeng X, Zhu S, Lu W, Liu Z, Huang J, Zhou Y, Fang J, Huang Y, Guo H, Li L, Trapp BD, Nussinov R, Eng C, Loscalzo J, Cheng F. Target identification among known drugs by deep learning from heterogeneous networks. Chem Sci. 2020;11(7):1775–97. https://doi.org/10.1039/C9SC04336E.
Kang D, Pang X, Lian W, Lvjie X, Wang J, Jia H, Zhang B, Liu AL, Guan-Hua D. Discovery of VEGFR2 inhibitors by integrating naïve Bayesian classification molecular docking and drug screening approaches. RSC Adv. 2018. https://doi.org/10.1039/c7ra12259d.
Al-Ali H, Lee D-H, Danzi MC, Nassif H, Gautam P, Wennerberg K, Zuercher B, Drewry DH, Lee JK, Lemmon VP, Bixby JL. Rational polypharmacology: systematically identifying and engaging multiple drug targets to promote axon growth. ACS Chem Biol. 2015;10(8):1939–51. https://doi.org/10.1021/acschembio.5b00289.
Huang YR, Bin YJ, Zeng PF, Lan W, Zhong C. NetPro: neighborhood interaction-based drug repositioning via label propagation. IEEE-ACM Trans Comput Biol Bioinform. 2023;20(3):2159–69. https://doi.org/10.1109/Tcbb.2023.3234331.
Lim H, Poleksic A, Yao Y, Tong H, He D, Zhuang L, Meng P, Xie L. Large-scale off-target identification using fast and accurate dual regularized one-class collaborative filtering and its application to drug repurposing. PLoS Comput Biol. 2016;12(10):e1005135. https://doi.org/10.1371/journal.pcbi.1005135.
Akdel M, Pires DEV, Pardo EP, Jänes J, Zalevsky AO, Mészáros B, Bryant P, Good LL, Laskowski RA, Pozzati G, Shenoy A, Zhu W, Kundrotas P, Serra VR, Rodrigues CHM, Dunham AS, Burke D, Borkakoti N, Velankar S, Frost A, Basquin J, Lindorff-Larsen K, Bateman A, Kajava AV, Valencia A, Ovchinnikov S, Durairaj J, Ascher DB, Thornton JM, Davey NE, Stein A, Elofsson A, Croll TI, Beltrao P. A structural biology community assessment of AlphaFold2 applications. Nat Struct Mol Biol. 2022;29(11):1056–67. https://doi.org/10.1038/s41594-022-00849-w.
Bordin N, Sillitoe I, Nallapareddy V, Rauer C, Lam SD, Waman VP, Sen N, Heinzinger M, Littmann M, Kim S, Velankar S, Steinegger M, Rost B, Orengo C. AlphaFold2 reveals commonalities and novelties in protein structure space for 21 model organisms. Commun Biol. 2023;6(1):1–12. https://doi.org/10.1038/s42003-023-04488-9.
Ye Q, Hsieh C-Y, Yang Z, Kang Y, Chen J, Cao D, He S, Hou T. A unified drug-target interaction prediction framework based on knowledge graph and recommendation system. Nat Commun. 2021;12(1):6775. https://doi.org/10.1038/s41467-021-27137-3.
Ahneman DT, Estrada JG, Lin S, Dreher SD, Doyle AG. Predicting reaction performance in C–N cross-coupling using machine learning. Science. 2018;360(6385):186–90. https://doi.org/10.1126/science.aar5169.
Steiner S, Wolf J, Glatzel S, Andreou A, Granda JM, Keenan G, Hinkley T, Aragon-Camarasa G, Kitson PJ, Angelone D, Cronin L. Organic synthesis in a modular robotic system driven by a chemical programming language. Science. 2019;363(6423):eaav2211. https://doi.org/10.1126/science.aav2211.
Xiong J, Xiong Z, Chen K, Jiang H, Zheng M. Graph neural networks for automated de novo drug design. Drug Discov Today. 2021;26(6):1382–93. https://doi.org/10.1016/j.drudis.2021.02.011.
Olivecrona M, Blaschke T, Engkvist O, Chen H. Molecular de-novo design through deep reinforcement learning. J Cheminf. 2017;9(1):1–14. https://doi.org/10.1186/s13321-017-0235-x.
Xiong G, Wu Z, Yi J, Fu L, Yang Z, Hsieh C, Yin M, Zeng X, Wu C, Lu A, Chen X, Hou T, Cao D. ADMETlab 2.0: an integrated online platform for accurate and comprehensive predictions of ADMET properties. Nucleic Acids Res. 2021. https://doi.org/10.1093/nar/gkab255.
Yang H, Lou C, Sun L, Li J, Cai Y, Wang Z, Li W, Liu G, Tang Y. admetSAR 2.0: web-service for prediction and optimization of chemical ADMET properties. Bioinformatics. 2019;35(6):1067–9. https://doi.org/10.1093/bioinformatics/bty707.
Ochoa D, Hercules A, Carmona M, Suveges D, Baker J, Malangone C, Lopez I, Miranda A, Cruz-Castillo C, Fumis L, Bernal-Llinares M, Tsukanov K, Cornu H, Tsirigos K, Razuvayevskaya O, Buniello A, Schwartzentruber J, Karim M, Ariano B, Osorio M, Ricardo E, Ferrer J, Ge X, Machlitt-Northen S, Gonzalez-Uriarte A, Saha S, Tirunagari S, Mehta C, Roldán-Romero JM, Horswell S, Young S, Ghoussaini M, Hulcoop DG, Dunham I, McDonagh EM. The next-generation open targets platform: reimagined, redesigned, rebuilt. Nucleic Acids Res. 2023. https://doi.org/10.1093/nar/gkac1046.
PandaOmics | Insilico Medicine.
Mkrtchyan GV, Veviorskiy A, Izumchenko E, Shneyderman A, Pun FW, Ozerov IV, Aliper A, Zhavoronkov A, Scheibye-Knudsen M. High-confidence cancer patient stratification through multiomics investigation of DNA repair disorders. Cell Death Dis. 2022;13(11):1–9. https://doi.org/10.1038/s41419-022-05437-w.
Ivanenkov YA, Polykovskiy D, Bezrukov D, Zagribelnyy B, Aladinskiy V, Kamya P, Aliper A, Ren F, Zhavoronkov A. Chemistry42: an ai-driven platform for molecular design and optimization. J Chem Inf Model. 2023;63(3):695–701. https://doi.org/10.1021/acs.jcim.2c01191.
McDermott J, Sturtevant D, Kathad U, Varma S, Zhou J, Kulkarni A, Biyani N, Schimke C, Reinhold WC, Elloumi F, Carr P, Pommier Y, Bhatia K. Artificial intelligence platform, RADR®, aids in the discovery of DNA damaging agent for the ultra-rare cancer atypical teratoid rhabdoid tumors. Front Drug Discov. 2022. https://doi.org/10.3389/fddsv.2022.1033395.
Lazarczyk M, Duda K, Mickael ME, Ak O, Paszkiewicz J, Kowalczyk A, Horbańczuk JO, Sacharczuk M. Adera2.0: a drug repurposing workflow for neuroimmunological investigations using neural networks. Molecules. 2022;27(19):6453. https://doi.org/10.3390/molecules27196453.
Sadeghi S, Lu J, Ngom A. A network-based drug repurposing method via non-negative matrix factorization. Bioinformatics. 2022;38(5):1369–77. https://doi.org/10.1093/bioinformatics/btab826.
Pavlopoulos GA, Paez-Espino D, Kyrpides NC, Iliopoulos I. Empirical comparison of visualization tools for larger-scale network analysis. Adv Bioinf. 2017;2017:e1278932. https://doi.org/10.1155/2017/1278932.
Xu B, Liu Y, Yu S, Wang L, Dong J, Lin H, Yang Z, Wang J, Xia F. A network embedding model for pathogenic genes prediction by multi-path random walking on heterogeneous network. BMC Med Genomics. 2019;12(S10):188. https://doi.org/10.1186/s12920-019-0627-z.
Weichselbraun A, Gindl S, Scharl A. Enriching semantic knowledge bases for opinion mining in big data applications. Knowledge-Based Syst. 2014;69:78–85. https://doi.org/10.1016/j.knosys.2014.04.039.
Ferrante M, Boccato T, Toschi N. Semantic brain decoding: from fMRI to conceptually similar image reconstruction of visual stimuli. [Preprint] 2023 Available from: https://doi.org/10.48550/arXiv.2212.06726
Domingo-Fernández D, Gadiya Y, Patel A, Mubeen S, Rivas-Barragan D, Diana CW, Misra BB, Healey D, Rokicki J, Colluru V. Causal reasoning over knowledge graphs leveraging drug-perturbed and disease-specific transcriptomic signatures for drug discovery. PLoS Comput Biol. 2022;18(2):e1009909. https://doi.org/10.1371/journal.pcbi.1009909.
Subpaiboonkit S, Li X, Zhao X, Scells H, Zuccon G. Causality discovery with domain knowledge for drug-drug interactions discovery. In: Li J, Wang S, Qin S, Li X, Wang S, editors. Advanced data mining and applications. Cham: Springer; 2019. p. 632–47.
Rivas-Barragan D, Mubeen S, Guim Bernat F, Hofmann-Apitius M, Domingo-Fernández D. Drug2ways: Reasoning over causal paths in biological networks for drug discovery. PLoS Comput Biol. 2020;16(12):e1008464. https://doi.org/10.1371/journal.pcbi.1008464.
Kondinski A, Bai J, Mosbach S, Akroyd J, Kraft M. Knowledge engineering in chemistry: from expert systems to agents of creation. Acc Chem Res. 2023;56(2):128–39. https://doi.org/10.1021/acs.accounts.2c00617.
Hasselgren A, Kralevska K, Gligoroski D, Pedersen SA, Faxvaag A. Blockchain in healthcare and health sciences—a scoping review. Int J Med Inf. 2020;134:104040. https://doi.org/10.1016/j.ijmedinf.2019.104040.
Haleem A, Javaid M, Singh RP, Suman R, Rab S. Blockchain technology applications in healthcare: an overview. Int J Intell Netw. 2021;2:130–9. https://doi.org/10.1016/j.ijin.2021.09.005.
Ghadge A, Bourlakis M, Kamble S, Seuring S. Blockchain implementation in pharmaceutical supply chains: A review and conceptual framework. Int J Prod Res. 2023;61(19):6633–51. https://doi.org/10.1080/00207543.2022.2125595.
Sarabi S, Han Q, de Vries B, Romme AGL, Almassy D. The nature-based solutions case-based system: a hybrid expert system. J Environ Manag. 2022;324:116413. https://doi.org/10.1016/j.jenvman.2022.116413.
Sundin I, Voronov A, Xiao H, Papadopoulos K, Bjerrum EJ, Heinonen M, Patronov A, Kaski S, Engkvist O. Human-in-the-loop assisted de novo molecular design. J Cheminf. 2022. https://doi.org/10.1186/s13321-022-00667-8.
Reker D. Practical considerations for active machine learning in drug discovery. Drug Discov Today: Technol. 2019. https://doi.org/10.1016/j.ddtec.2020.06.001.
Eisenstein M. Active machine learning helps drug hunters tackle biology. Nat Biotechnol. 2020;38(5):512–4. https://doi.org/10.1038/s41587-020-0521-4.
Ding X, Cui R, Yu J, Liu T, Zhu T, Wang D, Chang J, Fan Z, Liu X, Chen K, Jiang H, Li X, Luo X, Zheng M. Active learning for drug design: a case study on the plasma exposure of orally administered drugs. J Med Chem. 2021;64(22):16838–53. https://doi.org/10.1021/acs.jmedchem.1c01683.
A breakthrough milestone in AI-powered drug discovery reached linking biology and chemistry with AI. https://insilico.com/blog/pcc.
The Process and Costs of Drug Development. 2022. https://ftloscience.com/process-costs-drug-development/.
Funding
This work was supported by the National Key R&D Program of China (2023YFC3502900), the 3-year Action Plan for Shanghai TCM Development and Inheritance Program [ZY(2021-2023)-0401] and the National Natural Science Foundation of China (82104521).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors have no competing interests to declare that are relevant to the content of this article.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Chen, H., Lu, D., Xiao, Z. et al. Comprehensive applications of the artificial intelligence technology in new drug research and development. Health Inf Sci Syst 12, 41 (2024). https://doi.org/10.1007/s13755-024-00300-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13755-024-00300-y