Abstract
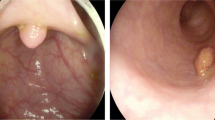
Colonoscopy screening test plays a crucial role in identifying and classifying possible cancerous polyps. The automation of polyp classification is challenging due to the limitations associated with the traditional handcrafted image features. This paper proposes a deep learning model for colorectal polyp classification, referred to as DeepCPC. This model consists of three main stages: image pre-processing using patch extraction and data augmentation; model initialization using six pre-trained CNNs to generate the fine-tuned baseline model; and formulating and learning generic yet discriminating image descriptors extracted and fused from the convolutional layers of two efficient CNNs architectures to classify colorectal polyps. The DeepCPC architecture has been fine-tuned on the CVC-Clinic dataset through a complete end-to-end training in which the patch extraction and image augmentation have been applied to generate more colonoscopy images for the patients. The experimental results show that the baseline fine-tuned model achieves an accuracy of 97.6%, and the final DeepCPC model achieves an accuracy of 98.4%. The reported results also demonstrate the capability of the proposed approach in identifying polyps in terms of precision, recall, and f-score. The DeepCPC helps the endoscopic physicians in classifying polyps and decreasing the colorectal polyp miss rate.









Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.References
Siegel, R.L., Miller, K.D., Goding Sauer, A., Fedewa, S.A., Butterly, L.F., Anderson, J.C., et al.: Colorectal cancer statistics. CA 70(3), 145–164 (2020)
Nair, M., Peate, I.: Pathophysiology for Nurses at a Glance. Wiley, New York (2015)
Groff, R.J., Nash, R., Ahnen, D.J.: Significance of serrated polyps of the colon. Curr. Gastroenterol. Rep. 10(5), 490–498 (2008)
Dekker, E., Rex, D.K.: Advances in CRC prevention: screening and surveillance. Gastroenterology 154(7), 1970–1984 (2018)
Arnold, M., Sierra, M.S., Laversanne, M., Soerjomataram, I., Jemal, A., Bray, F.: Global patterns and trends in colorectal cancer incidence and mortality. Gut 66(4), 683–691 (2017)
Imperiale, T.F., Ransohoff, D.F., Itzkowitz, S.H., Levin, T.R., Lavin, P., Lidgard, G.P., et al.: Multitarget stool DNA testing for colorectal-cancer screening. N. Engl. J. Med. 370(14), 1287–1297 (2014)
Barancin, C., Pickhardt, P.J., Kim, D.H., Spier, B., Lindstrom, M., Reichelderfer, M., et al.: Prospective blinded comparison of polyp size on computed tomography colonography and endoscopic colonoscopy. Clin. Gastroenterol. Hepatol. 9(5), 443–445 (2011)
Mir, A., Nguyen, V.Q., Soliman, Y., Sorrentino, D.: Wireless capsule endoscopy for diagnosis and management of post-operative recurrence of crohn’s disease. Life 11(7), 602 (2021)
Zauber, A.G., Winawer, S.J., O’Brien, M.J., Lansdorp-Vogelaar, I., van Ballegooijen, M., Hankey, B.F., et al.: Colonoscopic polypectomy and long-term prevention of colorectal-cancer deaths. N. Engl. J. Med. 366, 687–696 (2012)
Lieberman, D.A., Rex, D.K., Winawer, S.J., Giardiello, F.M., Johnson, D.A., Levin, T.R.: Guidelines for colonoscopy surveillance after screening and polypectomy: a consensus update by the US Multi-Society Task Force on Colorectal Cancer. Gastroenterology 143(3), 844–857 (2012)
Rex, D.K., Cutler, C.S., Lemmel, G.T., Rahmani, E.Y., Clark, D.W., Helper, D.J., et al.: Colonoscopic miss rates of adenomas determined by back-to-back colonoscopies. Gastroenterology 112(1), 24–28 (1997)
Kahi, C.J.: How does the serrated polyp pathway alter CRC screening and surveillance? Dig. Dis. Sci. 60(3), 773–780 (2015)
Tholoor, S., Tsagkournis, O., Basford, P., Bhandari, P.: Managing difficult polyps: techniques and pitfalls. Ann. Gastroenterol. 26(2), 114 (2013)
Irshad, H., Veillard, A., Roux, L., Racoceanu, D.: Methods for nuclei detection, segmentation, and classification in digital histopathology: a review—current status and future potential. IEEE Rev. Biomed. Eng. 7, 97–114 (2013)
Horváth, A., Spindler, S., Szalay, M., Rácz, I.: Preprocessing endoscopic images of colorectal polyps. Acta Technica Jaurinensis 9(1), 65–82 (2016)
Srivastava, A.N., Han, J. (eds.): Machine Learning and Knowledge Discovery for Engineering Systems Health Management. CRC Press, Boca Raton (2016)
Alzu’bi, A., Abuarqoub, A.: Deep learning model with low-dimensional random projection for large-scale image search. Eng. Sci. Technol. Int. J. 23(4), 911–920 (2020)
Jerebko, A.K., Malley, J.D., Franaszek, M., Summers, R.M.: Support vector machines committee classification method for computer-aided polyp detection in CT colonography1. Acad. Radiol. 12(4), 479–486 (2005)
Summers, R.M., Yao, J., Pickhardt, P.J., Franaszek, M., Bitter, I., Brickman, D., et al.: Computed tomographic virtual colonoscopy computer-aided polyp detection in a screening population. Gastroenterology 129(6), 1832–1844 (2005)
Mori, Y., Kudo, S.E., Berzin, T.M., Misawa, M., Takeda, K.: Computer-aided diagnosis for colonoscopy. Endoscopy 49(08), 813–819 (2017)
Ameling, S., Wirth, S., Paulus, D., Lacey, G., Vilarino, F.: Texture-based polyp detection in colonoscopy. In: Bildverarbeitung für die Medizin, pp. 346–350. Springer, Berlin (2009)
Karkanis, S.A., Iakovidis, D.K., Maroulis, D.E., Karras, D.A., Tzivras, M.: Computer-aided tumor detection in endoscopic video using color wavelet features. IEEE Trans. Inf. Technol. Biomed. 7(3), 141–152 (2003)
Pogorelov, K., Suman, S., Azmadi Hussin, F., Saeed Malik, A., Ostroukhova, O., Riegler, M., et al.: Bleeding detection in wireless capsule endoscopy videos—color versus texture features. J. Appl. Clin. Med. Phys. 20(8), 141–154 (2019)
Nogueira-Rodríguez, A., Domínguez-Carbajales, R., López-Fernández, H., Iglesias, Á., Cubiella, J., Fdez-Riverola, F., et al.: Deep neural networks approaches for detecting and classifying colorectal polyps. Neurocomputing 423, 721–734 (2021)
Obuch, J.C., Pigott, C.M., Ahnen, D.J.: Sessile serrated polyps: detection, eradication, and prevention of the evil twin. Curr. Treat. Options Gastroenterol. 13(1), 156–170 (2015)
Mesejo, P., Pizarro, D., Abergel, A., Rouquette, O., Beorchia, S., Poincloux, L., Bartoli, A.: Computer-aided classification of gastrointestinal lesions in regular colonoscopy. IEEE Trans. Med. Imaging 35(9), 2051–2063 (2016)
Tamaki, T., Yoshimuta, J., Kawakami, M., Raytchev, B., Kaneda, K., Yoshida, S., et al.: Computer-aided colorectal tumor classification in NBI endoscopy using local features. Med. Image Anal. 17(1), 78–100 (2013)
Stehle, T., Auer, R., Gross, S., Behrens, A., Wulff, J., Aach, T., et al.: Classification of colon polyps in NBI endoscopy using vascularization features. In: Medical Imaging 2009: Computer-Aided Diagnosis, International Society for Optics and Photonics, 7260, 72602S (2009)
Li, P., Chan, K.L., Krishnan, S.M., Gao, Y.: Detecting abnormal regions in colonoscopic images by patch-based classifier ensemble. In: Proceedings of the 17th International Conference on Pattern Recognition, vol. 3, pp. 774–777 (2004)
Manivannan, S., Wang, R., Trucco, E., Hood, A.: Automatic normal-abnormal video frame classification for colonoscopy. In: 2013 IEEE 10th International Symposium on Biomedical Imaging, pp. 644–647 (2013)
Hilsden, R.J., Heitman, S.J., Mizrahi, B., Narod, S.A., Goshen, R.: Prediction of findings at screening colonoscopy using a machine learning algorithm based on complete blood counts (ColonFlag). PLoS ONE 13(11), e0207848 (2018)
Ribeiro, E., Uhl, A., Häfner, M.: Colonic polyp classification with convolutional neural networks. In: 2016 IEEE 29th International Symposium on Computer-Based Medical Systems (CBMS), pp. 253–258 (2016)
Byrne, M.F., Chapados, N., Soudan, F., Oertel, C., Pérez, M.L., Kelly, R., et al.: Real-time differentiation of adenomatous and hyperplastic diminutive colorectal polyps during analysis of unaltered videos of standard colonoscopy using a deep learning model. Gut 68(1), 94–100 (2019)
Tian, Y., Pu, L. Z., Singh, R., Burt, A. D., Carneiro, G.: One-stage five-class polyp detection and classification. In: 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019), pp. 70–73 (2019)
Komeda, Y., Handa, H., Matsui, R., Kashida, H., Watanabe, T., Sakurai, T., Kudo, M.: Computer-aided diagnosis (CAD) based on convolutional neural network (CNN) system using artificial intelligence (AI) for colorectal polyp classification. Endoscopy 51(04), OP2 (2019)
Wang, L., Chen, R., Wang, S., Zeng, N., Huang, X., Liu, C.: Nested dilation network (NDN) for multi-task medical image segmentation. IEEE Access 7, 44676–44685 (2019)
Wimmer, G., Vécsei, A., Uhl, A.: CNN transfer learning for the automated diagnosis of celiac disease. In: 2016 Sixth International Conference on Image Processing Theory, Tools and Applications (IPTA), pp. 1–6 (2016)
Urban, G., Tripathi, P., Alkayali, T., Mittal, M., Jalali, F., Karnes, W., Baldi, P.: Deep learning localizes and identifies polyps in real time with 96% accuracy in screening colonoscopy. Gastroenterology 155(4), 1069–1078 (2018)
Patino-Barrientos, S., Sierra-Sosa, D., Garcia-Zapirain, B., Castillo-Olea, C., Elmaghraby, A.: Kudo’s classification for colon polyps assessment using a deep learning approach. Appl. Sci. 10(2), 501 (2020)
Patel, K., Li, K., Tao, K., Wang, Q., Bansal, A., Rastogi, A., Wang, G.: A comparative study on polyp classification using convolutional neural networks. PLoS ONE 15(7), e0236452 (2020)
Alzu’bi, A., Amira, A., Ramzan, N.: Learning transfer using deep convolutional features for remote sensing image retrieval. Int. J. Comput. Sci. 46(4), 637–644 (2019)
Weiss, K., Khoshgoftaar, T.M., Wang, D.: A survey of transfer learning. J. Big data 3(1), 1–40 (2016)
Deng, J., Dong, W., Socher, R., Li, L.J., Li, K., Fei-Fei, L.: Imagenet: a large-scale hierarchical image database. In: 2009 IEEE Conference on Computer Vision and Pattern Recognition, pp. 248–255 (2009)
Bernal, J., Sánchez, F.J., Fernández-Esparrach, G., Gil, D., Rodríguez, C., Vilariño, F.: WM-DOVA maps for accurate polyp highlighting in colonoscopy: validation vs. saliency maps from physicians. Comput. Med. Imaging Graphics 43, 99–111 (2015)
He, K., Zhang, X., Ren, S., Sun, J.: Deep residual learning for image recognition. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 770–778 (2016)
Simonyan, K., Zisserman, A.: Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:abs/1409.1556 (2014)
Szegedy, C., Liu, W., Jia, Y., Sermanet, P., Reed, S., Anguelov, D., et al.: Going deeper with convolutions. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 1–9 (2015)
Sandler, M., Howard, A., Zhu, M., Zhmoginov, A., Chen, L.C.: Mobilenetv2: inverted residuals and linear bottlenecks. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 4510–4520 (2018)
Chollet, F.: Xception: deep learning with depthwise separable convolutions. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 1251–1258 (2017)
Bisong, E.: Building machine learning and deep learning models on Google cloud platform: a comprehensive guide for beginners. Apress. 7–10 (2019)
Huang, Y., Gong, W., Su, B., Zhi, F., Liu, S., Jiang, B.: Risk and cause of interval colorectal cancer after colonoscopic polypectomy. Digestion 86(2), 148–154
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Taha, D., Alzu’bi, A., Abuarqoub, A. et al. Automated Colorectal Polyp Classification Using Deep Neural Networks with Colonoscopy Images. Int. J. Fuzzy Syst. 24, 2525–2537 (2022). https://doi.org/10.1007/s40815-021-01182-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40815-021-01182-y