Abstract
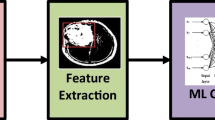
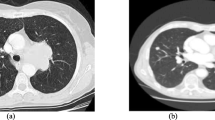
Lung cancer is the type of cancer that causes the most deaths each year. It is also cancer with the lowest survival rate. This represents a health problem worldwide. Lung cancer has two subtypes: Non-Small Cell Lung Cancer (NSCLC) and Small Cell Lung Cancer (SCLC). For doctors, it can be hard to detect and differentiate them. Therefore, in this work, we present a method to help doctors with this issue. It consists of three phases: image preprocessing is the first phase. It starts gathering the data. After that, PET scans are selected. Then, all the scans are converted to grayscale images, and finally, all the images are joined to create a video from each patient’s scan. Next, the data extraction phase starts. In this phase, some frames are extracted from each video, and they are flattened and blended to create a row of information from each frame. Thus, a dataframe is created where each row represents a patient, and each column is a pixel value. To obtain better results, an oversampling technique is applied. In this manner, the classes are balanced. Following this, a dimensionality reduction technique is applied to reduce the number of columns produced by the previous steps and to check if this technique improves the results yielded by each model. Subsequently, the model evaluation phase begins. At this stage, two models are created: a Support Vector Machine (SVM), and a Random Forest. Ultimately, the findings are unveiled, revealing that the SVM emerged as the top-performing model, boasting an impressive 97% accuracy, 98% precision, and 97% sensitivity. Eventually, this method can be applied to detect and classify different diseases that involve PET scans.

Similar content being viewed by others
Data availability
The dataset utilized in this research is sourced from "The Cancer Imaging Archive (TCIA)," accessible at https://imaging.cancer.gov/informatics/cancer_imaging_archive.htm. TCIA provides a comprehensive collection of openly available medical images, fostering collaborative research in cancer imaging.
Code Availability
The code implemented in this study is openly available on the following URL: https://colab.research.google.com/drive/1JKq3NV4wCkN1tTbwVkU-bNmL4qt2NPpy?usp=sharing. Researchers interested in reproducing or building upon the methods employed in this paper are encouraged to access the provided code. This transparency aims to facilitate the reproducibility of our findings and promote collaborative efforts in advancing cancer imaging research.
Notes
More information can be found in: https://keras.io/examples/vision/video_classification/.
For more information refer to: https://imbalanced-learn.org/stable/references/generated/imblearn.over_sampling.SMOTE.html.
References
Anowar F, Sadaoui S, Selim B. Conceptual and empirical comparison of dimensionality reduction algorithms (pca, kpca, lda, mds, svd, lle, isomap, le, ica, t-sne). Comput Sci Rev. 2021;40:100378. https://www.sciencedirect.com/science/article/pii/S1574013721000186. Accessed 13 Dec 2022
Bade BC, Dela Cruz CS. Lung cancer 2020: epidemiology, etiology, and prevention. Clin Chest Med. 2020;41(1):1–24. https://doi.org/10.1016/j.ccm.2019.10.001.
Barta JA, Powell CA, Wisnivesky JP. Global epidemiology of lung cancer. Ann Glob Health. 2019;85(1):8–24
Biau G, Scornet E. A random forest guided tour. TEST. 2016;25(2):197–227.
Cano JR, Gutiérrez PA, Krawczyk B, Woźniak M, García S. Monotonic classification: an overview on algorithms, performance measures and data sets. Neurocomputing. 2019;341:168–82.
Chauhan VK, Dahiya K, Sharma A. Problem formulations and solvers in linear svm: a review. Artif Intell Rev. 2019;52(2):803–55.
Chen CC, Li ST. Credit rating with a monotonicity-constrained support vector machine model. Expert Syst Appl. 2014;41(16):7235–47.
Clark K, Vendt B, Smith K, Freymann J, Kirby J, Koppel P, Moore S, Phillips S, Maffitt D, Pringle M, Tarbox L, Prior F. The cancer imaging archive (TCIA): maintaining and operating a public information repository. J Digit Imaging. 2013;26(6):1045–57.
Howlader N, Forjaz G, Mooradian MJ, Meza R, Kong CY, Cronin KA, Mariotto AB, Lowy DR, Feuer EJ. The effect of advances in lung-cancer treatment on population mortality. N Engl J Med. 2020;383(7):640–9. https://doi.org/10.1056/NEJMoa1916623. (pMID: 32786189).
Huang H, Zheng D, Chen H, Wang Y, Chen C, Xu L, Li G, Wang Y, He X, Li W. Fusion of ct images and clinical variables based on deep learning for predicting invasiveness risk of stage i lung adenocarcinoma. Med Phys. 2022;49(10):6384–94. https://doi.org/10.1002/mp.15903.
Huang S, Cai N, Pacheco PP, Narrandes S, Wang Y, Xu W. Applications of support vector machine (svm) learning in cancer genomics. Cancer Genom Proteom. 2018;15(1):41–51.
Lameka K, Farwell MD, Ichise M. Chapter 11 - positron emission tomography. In: Masdeu, JC, González RG. editors. Neuroimaging part I, handbook of clinical neurology, vol. 135. Elsevier; 2016, p. 209–227. Elsevier. https://www.sciencedirect.com/science/article/pii/B9780444534859000118.
Li P, Wang S, Li T, Lu J, HuangFu Y, Wang D. A large-scale CT and PET/CT dataset for lung cancer diagnosis. 2020. https://doi.org/10.7937/TCIA.2020.NNC2-0461.
Liashchynskyi P, Liashchynskyi P. Grid search, random search, genetic algorithm: a big comparison for nas. 2019.
Ma Y, Feng W, Wu Z, Liu M, Zhang F, Liang Z, Cui C, Huang J, Li X, Guo X. Intra-tumoural heterogeneity characterization through texture and colour analysis for differentiation of non-small cell lung carcinoma subtypes. Phys Med Biol. 2018;63(16): 165018.
Makaju S, Prasad P, Alsadoon A, Singh A, Elchouemi A. Lung cancer detection using ct scan images. Proc Comput Sci. 2018;125:107–14. In: The 6th International Conference on smart computing and communications. https://www.sciencedirect.com/science/article/pii/S1877050917327801. Accessed 19 Dec 2022
Nooreldeen R, Bach H. Current and future development in lung cancer diagnosis. Int J Mol Sci. 2021;22(16). https://www.mdpi.com/1422-0067/22/16/8661. Accessed 7 Jan 2023
Park YJ, Choi D, Choi JY, Hyun SH. Performance evaluation of a deep learning system for differential diagnosis of lung cancer with conventional ct and fdg pet/ct using transfer learning and metadata. Clin Nucl Med. 2021;46(8):635–40.
Saito T, Rehmsmeier M. The precision-recall plot is more informative than the roc plot when evaluating binary classifiers on imbalanced datasets. PLoS One. 2015;10(3):1–21.
Schabath MB, Cote ML. Cancer progress and priorities: lung cancer. Cancer Epidemiol Biomark Prev. 2019;28(10):1563–79. https://doi.org/10.1158/1055-9965.EPI-19-0221.
Soltanzadeh P, Hashemzadeh M. Rcsmote: range-controlled synthetic minority over-sampling technique for handling the class imbalance problem. Inform Sci. 2021;542:92–111. https://www.sciencedirect.com/science/article/pii/S0020025520306794. Accessed 15 Dec 2022
Tam M, Dyer T, Dissez G, Morgan TN, Hughes M, Illes J, Rasalingham R, Rasalingham S. Augmenting lung cancer diagnosis on chest radiographs: positioning artificial intelligence to improve radiologist performance. Clin Radiol. 2021;76(8):607–14. https://www.sciencedirect.com/science/article/pii/S0009926021002373. Accessed 3 Jan 2023
Tanoue LT, Tanner NT, Gould MK, Silvestri GA. Lung cancer screening. Am J Respir Crit Care Med. 2015;191(1):19–33.
Thai AA, Solomon BJ, Sequist LV, Gainor JF, Heist RS. Lung cancer. Lancet. 2021;398(10299):535–54. https://doi.org/10.1016/S0140-6736(21)00312-3.
Wood DE, Eapen GA, Ettinger DS, Hou L, Jackman D, Kazerooni E, Klippenstein D, Lackner RP, Leard L, Leung AN, et al. Lung cancer screening. J Natl Compr Canc Netw. 2012;10(2):240–65.
Yin Z, Hou J. Recent advances on svm based fault diagnosis and process monitoring in complicated industrial processes. Neurocomputing. 2016;174:643–50. https://www.sciencedirect.com/science/article/pii/S0925231215014149. Accessed 15 Dec 2022
Acknowledgements
This research has been supported by the “Sistemas Inteligentes de Soporte a la Educación Especial (SINSAE v5)” research project of the UNESCO Chair on Support Technologies for Educational Inclusion.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors of this paper declare that they have no conflicts of interest that could potentially influence or bias the interpretation of the research presented. There are no financial, personal, or professional relationships that might be perceived as having influenced the work reported in this manuscript. This includes, but is not limited to, financial interests, affiliations, or relationships with organizations that may have a direct or indirect interest in the subject matter discussed in the paper. We affirm that this work is conducted with integrity and in compliance with ethical standards. Any external factors that could pose a conflict of interest have been disclosed in an honest and transparent manner.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the topical collection “Emerging Technologies in Applied Informatics” guest edited by Hector Florez and Marcelo Leon.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Jara-Gavilanes, A., Robles-Bykbaev, V. Lung Cancer Detection: A Classification Approach Utilizing Oversampling and Support Vector Machines. SN COMPUT. SCI. 5, 74 (2024). https://doi.org/10.1007/s42979-023-02432-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s42979-023-02432-6