Abstract
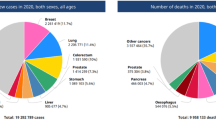
The human behaviors are varying depends on the DNA module presents in generation of human life styles. Increasing disease factors in due to DNA from history of family members continuously affects the present’s life. In the sense, the Colon Cancer (CC) is one of the deadliest diseases having the damaged cells to affect the human gene cells to damage cause deaths. The most case limitations uncovered from existing level is non mutual feature selection due to non-impact feature margin decrease the precision and identification accuracy. To resolve this problem, we propose a genetics sequence linear support vector (GSLSV) based feature selection with adaptive ensemble boosting fuzzified deep neural network (AEB-FDNN) to identify the colon cancer. Initially the preprocessing is carried by Box-plot normalization process (Bp-Np) to make noise less transformation. Then synthetic minority oversampling technique (SMOsT) is applied to identify the mutual relational features weight which is support for identifying cancer impact margins. Then colon cancer periodic influence rate (CCPIR) is applied to find the disease affection rate. With the support of marginal weights, the GSLSV based feature selection is applied to select the relation mutual features. Then the selected features is applied using AEB-FDNN to identify the CC based on the risk by class. The proposed system effectively train the dataset by improving the testing and validating accuracy as well higher performance in precision, recall rate, f1-measure and identification accuracy compared to the other system.









Similar content being viewed by others
Data Availability
The dataset generated and analyzed during the current study are available from the corresponding author on reasonable request.
References
Shafi ASM, Molla MMI, Jui JJ, et al. Detection of colon cancer based on microarray dataset using machine learning as a feature selection and classification techniques. SN Appl Sci. 2020;2:1243. https://doi.org/10.1007/s42452-020-3051-2.
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424.
Xu H, Ma Y, Zhang J, Gu J, Jing X, Lu S, Fu S, Huo J. Identification and verification of core genes in colorectal cancer. BioMed Res Int. 2020;2020:8082697. https://doi.org/10.1155/2020/8082697.
Breugom AJ, et al. Adjuvant chemotherapy and relative survival of patients with stage II colon cancer-A EURECCA international comparison between the Netherlands, Denmark, Sweden, England, Ireland, Belgium, and Lithuania. Eur J Cancer. 2016;63:110–7.
Yu H, Ni J, Dan Y, Xu S. Mining and integrating reliable decision rules for imbalanced cancer gene expression data sets. Tsinghua Sci Technol. 2012;17(6):666–73. https://doi.org/10.1109/TST.2012.6374368.
Rathore S, Hussain M, Ali A, Khan A. A recent survey on colon cancer detection techniques. IEEE/ACM Trans Comput Biol Bioinform. 2013;10(3):545–63. https://doi.org/10.1109/TCBB.2013.84.
Rathore S, Hussain M, Khan A. GECC: gene expression based ensemble classification of colon samples. IEEE/ACM Trans Comput Biol Bioinform. 2014;11(6):1131–45. https://doi.org/10.1109/TCBB.2014.2344655.
Xu T, Ou-Yang L, Hu X, Zhang X-F. Identifying gene network rewiring by integrating gene expression and gene network data. IEEE/ACM Trans Comput Biol Bioinform. 2018;15(6):2079–85. https://doi.org/10.1109/TCBB.2018.2809603.
Li J, Dong W, Meng D. Grouped gene selection of cancer via adaptive sparse group lasso based on conditional mutual information. IEEE/ACM Trans Comput Biol Bioinform. 2018;15(6):2028–38. https://doi.org/10.1109/TCBB.2017.2761871.
Liu K, Ye J, Yang Y, Shen L, Jiang H. A unified model for joint normalization and differential gene expression detection in RNA-Seq data. IEEE/ACM Trans Comput Biol Bioinform. 2019;16(2):442–54. https://doi.org/10.1109/TCBB.2018.2790918.
Paul S, Brahma D. An integrated approach for identification of functionally similar microRNAs in colorectal cancer. IEEE/ACM Trans Comput Biol Bioinform. 2019;16(1):183–92. https://doi.org/10.1109/TCBB.2017.2765332.
Liu J-X, Feng C-M, Kong X-Z, Xu Y. Dual graph-Laplacian PCA: a closed-form solution for bi-clustering to find “Checkerboard” structures on gene expression data. IEEE Access. 2019;7:151329–38. https://doi.org/10.1109/ACCESS.2019.2941227.
Waseem MH, et al. On the feature selection methods and reject option classifiers for robust cancer prediction. IEEE Access. 2019;7:141072–82. https://doi.org/10.1109/ACCESS.2019.2944295.
Maji P. f -information measures for efficient selection of discriminative genes from microarray data. IEEE Trans Biomed Eng. 2009;56(4):1063–9. https://doi.org/10.1109/TBME.2008.2004502.
Chan SC, Wu HC, Tsui KM. A new method for preliminary identification of gene regulatory networks from gene microarray cancer data using ridge partial least squares with recursive feature elimination and novel brier and occurrence probability measures. IEEE Trans Syst Man Cybern A Syst Hum. 2012;42(6):1514–28. https://doi.org/10.1109/TSMCA.2012.2199302.
Tahir A, et al. Survivability period prediction in colon cancer patients using machine learning. In: 2023 International conference on energy, power, environment, control, and computing (ICEPECC), Gujrat, Pakistan. 2023. p. 1–4. https://doi.org/10.1109/ICEPECC57281.2023.10209530
Zhao X, Yang Y, Yin M. MHRWR: prediction of lncRNA-disease associations based on multiple heterogeneous networks. IEEE/ACM Trans Comput Biol Bioinform. 2021;18(6):2577–85. https://doi.org/10.1109/TCBB.2020.2974732.
Horaira MA, Ahmed MS, Kabir MH, Mollah MNH, Rahman Shah MA. Colon cancer prediction from gene expression profiles using kernel based support vector machine. In: 2018 International conference on computer, communication, chemical, material and electronic engineering (IC4ME2), Rajshahi, Bangladesh. 2018. p. 1–4.https://doi.org/10.1109/IC4ME2.2018.8465636
Saheed YK, Balogun BF, Odunayo BJ, Abdulsalam M. Microarray gene expression data classification via Wilcoxon sign rank sum and novel Grey Wolf optimized ensemble learning models. IEEE/ACM Trans Comput Biol Bioinform. 2023;20(6):3575–87. https://doi.org/10.1109/TCBB.2023.3305429.
Thakur T, Batra I, Malik A, Ghimire D, Kim S-H, Sanwar Hosen ASM. RNN-CNN based cancer prediction model for gene expression. IEEE Access. 2023;11:131024–44. https://doi.org/10.1109/ACCESS.2023.3332479.
Li S, Yang Y, Wang X, et al. Colorectal cancer subtype identification from differential gene expression levels using minimalist deep learning. BioData Min. 2022;15:12. https://doi.org/10.1186/s13040-022-00295-w.
Niu L, Gao C, Li Y. Identification of potential core genes in colorectal carcinoma and key genes in colorectal cancer liver metastasis using bioinformatics analysis. Sci Rep. 2021;11:23938. https://doi.org/10.1038/s41598-021-03395-5.
Gupta S, Gupta MK, Shabaz M, Sharma A. Deep learning techniques for cancer classification using microarray gene expression data. Front Physiol. 2022;13:952709. https://doi.org/10.3389/fphys.2022.952709.
Al-Rajab M, Lu J, Xu Q. A framework model using multifilter feature selection to enhance colon cancer classification. PLoS ONE. 2021;16(4):e0249094. https://doi.org/10.1371/journal.pone.0249094.
Salimy S, Lanjanian H, et al. A deep learning-based framework for predicting survival-associated groups in colon cancer by integrating multi-omics and clinical data. Heliyon. 2023;9:1–16.
Muazzam F. Multi-class cancer classification and biomarker identification using deep learning. National University of Computer And Emerging Sciences. 2021. p.1–12.
Acknowledgements
The authors acknowledged the Thanthai Periyar Govt. Arts & Science College (A), Tiruchirappalli, India for supporting the research work by providing the facilities.
Funding
No funding received for this research.
Author information
Authors and Affiliations
Contributions
This research endeavor was made possible by the collaboration and contributions of all authors.
Corresponding author
Ethics declarations
Conflict of interest
No conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the topical collection “Advances in Computational Approaches for Image Processing, Wireless Networks, Cloud Applications and Network Security” guest edited by P. Raviraj, Maode Ma and Roopashree H R.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Benazir Butto, S., FathimaBibi, K. Efficient Colon Cancer Identification Based on Genetics Sequence Linear Support Vector Feature Selection Using Adaptive Ensemble Boosting Fuzzified Deep Neural Network. SN COMPUT. SCI. 5, 601 (2024). https://doi.org/10.1007/s42979-024-02925-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s42979-024-02925-y