Abstract
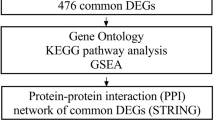
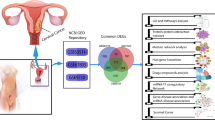
Cervical cancer is a threat to women worldwide, with poor early prognosis. Cervical cancer is strongly linked to human papillomavirus (HPV), particularly high-risk HPV16 and HPV18, a prevalent gynecological malignancy. It ranks fourth among women's malignant diseases in terms of morbidity and mortality and there has been a rise in the incidence of cervical cancer among young females in recent decades. The Gene Expression Omnibus database (GEO) was used to screen and download three gene expression profiles (GSE63514, GSE6791, and GSE9750). The study used GEO2R and Venn diagrams to identify differentially expressed genes (DEGs) across three gene expression datasets. GO and KEGG Pathway Enrichment Analysis (PEA) were then performed on the DEGs to understand their biological functions and signaling pathways. Gene set enrichment analysis (GSEA) is also applied to analyze expression profiles. Additionally, a protein–protein interaction (PPI) network is constructed using the DEGs, which underwent a functionality-based enrichment investigation to identify hub genes. The proposed approach has clinical relevance in uncovering potential therapeutic targets and improving our understanding of the prognosis and treatment of the disease.





Similar content being viewed by others
Data Availability
The dataset produced and scrutinized in this study are accessible from the corresponding author upon reasonable request.
References
Yang HJ, Xue JM, Li J, Wan LH, Zhu YX. Identification of key genes and pathways of diagnosis and prognosis in cervical cancer by bioinformatics analysis. Mol Genet Genomic Med. 2020;8(6): e1200. https://doi.org/10.1002/mgg3.1200.
Ali A, Ajil A, Meenakshi Sundaram A, Joseph N. Detection of gene ontology clusters using biclustering algorithms. SN Comput Sci. 2023;4(3):217. https://doi.org/10.1007/s42979-022-01624-w.
Elias MH, Das S, Abdul Hamid N. Candidate genes and pathways in cervical cancer: a systematic review and integrated bioinformatic analysis. Cancers. 2023;15(3):853.
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. https://doi.org/10.3322/caac.21492.
Kessler TA. Cervical cancer: prevention and early detection. In Seminars in oncology nursing, vol. 33, No. 2, WB Saunders. 2017. pp. 172–183. https://doi.org/10.1016/j.soncn.2017.02.005
Ali A, et al. DPEBic: detecting essential proteins in gene expressions using encoding and biclustering algorithm. J Ambient Intell Humaniz Comput. 2021. https://doi.org/10.1007/s12652-021-03036-9.
Hasan MT, Islam MR, Islam MR, Altahan BR, Ahmed K, Bui FM, Moni MA. Systematic approach to identify therapeutic targets and functional pathways for the cervical cancer. J Genet Eng Biotechnol. 2023;21(1):10. https://doi.org/10.1186/s43141-023-00469-x.
Ali A. et al. A review of aligners for protein protein interaction networks. In 2017 2nd IEEE international conference on recent trends in electronics, information and communication technology (RTEICT). IEEE, 2017. https://doi.org/10.1109/RTEICT.2017.8256879
Andalib KS, Rahman MH, Habib A. Bioinformatics and cheminformatics approaches to identify pathways, molecular mechanisms and drug substances related to genetic basis of cervical cancer. J Biomol Struct Dyn. 2023;41(23):14232–47. https://doi.org/10.1080/07391102.2023.2179542.
den Boon JA, Pyeon D, Wang SS, Horswill M, Schiffman M, Sherman M, Ahlquist P. Molecular transitions from papillomavirus infection to cervical precancer and cancer: role of stromal estrogen receptor signaling. Proc Natl Acad Sci U S A. 2015;112(25):E3255–64. https://doi.org/10.1073/pnas.1509322112.
Pyeon D, Newton MA, Lambert PF, den Boon JA, Sengupta S, Marsit CJ, Ahlquist P. Fundamental differences in cell cycle deregulation in human papillomavirus-positive and human papillomavirus-negative head/neck and cervical cancers. Cancer Res. 2007;67(10):4605–19. https://doi.org/10.1158/0008-5472.Can-06-3619.
Ali A, Hulipalled VR, Patil SS, Adbulkader R. Consensus pattern selection from structured profile using multiobjective algorithm. Int J Adv Sci Technol. 2019;28(8):294–305.
Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Soboleva A. NCBI GEO: archive for functional genomics data sets—update. Nucl Acids Res. 2012;41(D1):D991–5. https://doi.org/10.1093/nar/gks1193.
Saga Y, Suzuki M, Mizukami H, Kohno T, Takei Y, Fukushima M, Ozawa K. Overexpression of thymidylate synthase mediates desensitization for 5-fluorouracil of tumor cells. Int J Cancer. 2003;106(3):324–6. https://doi.org/10.1002/ijc.11221.
Ramachandra HV, Ali A, Ambili PS, Thota S, Asha PN. An optimization on bicluster algorithm for gene expression data. In 2023 4th IEEE global conference for advancement in technology (GCAT), 2023. pp. 1–6. IEEE. https://doi.org/10.1109/GCAT59970.2023.10353373
Ali A, Ramachandra HV, Meenakshi Sundaram A, Ajil A, Ramakrishnan N. Pareto optimization technique for protein motif detection in genomic data set. In International Conference on Information, Communication and Computing Technology, Singapore: Springer Nature Singapore, 2023. pp. 963–977. https://doi.org/10.1007/978-981-99-5166-6_65
Khan M, Hameed Y. Discovery of novel six genes-based cervical cancer-associated biomarkers that are capable to break the heterogeneity barrier and applicable at the global level. J Cancer Res Ther. 2023. https://doi.org/10.4103/jcrt.jcrt_1588_21.
Ali A, Hulipalled VR, Patil SS. Centrality measure analysis on protein interaction networks. In 2020 IEEE international conference on technology, engineering, management for societal impact using marketing, entrepreneurship and talent (TEMSMET). IEEE, 2020. https://doi.org/10.1109/TEMSMET51618.2020.9557447
Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, et al. “STRI NG v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucl Acids Res. 2019;47(D1):D607-d613. https://doi.org/10.1093/nar/gky1131.
Razia S, Nakayama K, Nakamura K, Ishibashi T, Ishikawa M, Minamoto T, Kyo S. Clinicopathological and biological analysis of PIK3CA mutation and amplification in cervical carcinomas. Exp Ther Med. 2019;18(3):2278–84. https://doi.org/10.3892/etm.2019.7771.
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–504. https://doi.org/10.1101/gr.1239303.
Shi H, Han X, Sun Y, Shang C, Wei M, Ba X, Zeng X. Chemokine (C-X-C motif) ligand 1 and CXCL2 produced by tumor promote the generation of monocytic myeloid-derived suppressor cells. Cancer Sci. 2018;109(12):3826–39. https://doi.org/10.1111/cas.13809.
Shukla V, Mallya S, Adiga D, Sriharikrishnaa S, Chakrabarty S, Kabekkodu SP. Bioinformatic analysis of miR-200b/429 and hub gene network in cervical cancer. Biochem Genet. 2023;61(5):1898–916. https://doi.org/10.1007/s10528-023-10356-2.
Kumar M, Ravishankar H, Deepa KR, et al. Early diagnosis of COVID-19 disease by chestnet convolutional neural network from chest Xray images. SN Comput Sci. 2024;5:696. https://doi.org/10.1007/s42979-024-02998-9.
Prem Kumar M, Ravi Shankar H, Deepa KR, et al. Effective COVID-19 disease identification using correlation coefficient absolute feature selection and logistic boosting neural network algorithm. SN Comput Sci. 2024;5:662. https://doi.org/10.1007/s42979-024-02941-y.
Huchappa R, Patil KK. Evolutionary model to guarantee quality of service for tactical worldwide interoperability for microwave access networks. IAES Int J Artif Intell. 2022;11(2):687.
Ravishankar H, Patil KK. Throughput optimized using evolutionary computing to guarantee QoS in IEEE 802.16 networks. In 2017 international conference on smart technologies for smart nation (SmartTechCon), Bengaluru, India, 2017. pp. 1602–1606, https://doi.org/10.1109/SmartTechCon.2017.8358635
Acknowledgements
The authors acknowledged the REVA University, Bangalore, Karnataka, India; Ilahia college of Engineering and Technology, Ernakulam, Kerala, India; Wipro Arabia Ltd., Dhahran, Saudi Arabia for supporting the research work by providing the facilities.
Funding
No funding received for this research.
Author information
Authors and Affiliations
Contributions
This research endeavor was made possible by the collaboration and contributions of all authors.
Corresponding author
Ethics declarations
Conflict of interest
No conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ali, A., Mohan, J., Nadaf, T.A.A. et al. Bioinformatics-Driven Discovery of Signaling Pathways and Genes Influencing Cervical Cancer. SN COMPUT. SCI. 5, 989 (2024). https://doi.org/10.1007/s42979-024-03347-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s42979-024-03347-6