Abstract
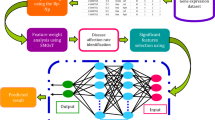
Many types of cancer are prevalent in humans today, and unfortunately, these cancers are responsible for the death of many people. Cancer is a lethal disease yielded by various genetic and biochemical abnormalities. In the world, Colon Cancer (CC) is the fourth most prevalent cancer to be diagnosed and the third greatest cause of mortality. Because there are no reliable diagnostic indicators and nothing is known about the underlying molecular pathways, the death rate from CC keeps rising. The existing procedure of CC detection is an error-prone and time-consuming process. Also, owing to improper features produced high misclassification. Different stages of cancer modify the expression levels of particular molecules, making early diagnosis and detection challenging. As a result, there is an increased urgency to develop accurate CC detection. To tackle this issue, this paper introduces the Genetic Decision Support Regression (GDSR) algorithm and Exhaustive Correlation Feature Selection (ECFS) for colon cancer detection. The first step involves collecting samples of the colon cancer dataset from the Kaggle repository and normalizing the entire dataset using the Box-plot Normalization process (Bp-Np) technique. The following phase uses the Synthetic Minority Oversampling Technique (SMOTE) approach to determine feature weight. The colon cancer affection rate is then examined using the CC Periodic Influence Rate (CCPIR) approach. Following this, the ECFS algorithm is applied to select suitable colon cancer attributes to reduce the dataset's dimensionality. Finally, the GDSR algorithm is utilized to classify colon cancer based on the best attributes. The proposed algorithm exhibits high classification accuracy, precision, and recall performance. It also reduces the time required for colon cancer detection and has a lower false rate compared to alternative methods.











Similar content being viewed by others
Data Availability
The dataset generated and analyzed during the current study are available from the corresponding author on reasonable request.
References
Fahami MA, Roshanzamir M, Izadi NH, Keyvani V, Alizadehsani R. Detection of effective genes in colon cancer: a machine learning approach. Inf Med Unlocked. 2021;24:100605.
Koppad S, Basava A, Nash K, Gkoutos GV, Acharjee A. Machine learning-based identification of colon cancer candidate diagnostics genes. Biology. 2022;11(3):365. https://doi.org/10.3390/biology11030365.
Maurya NS, Kushwaha S, Chawade A, et al. Transcriptome profiling by combined machine learning and statistical R analysis identifies TMEM236 as a potential novel diagnostic biomarker for colorectal cancer. Sci Rep. 2021;11:14304.
Paksoy N, Yağın FH. Artificial intelligence-based colon cancer prediction by identifying genomic biomarkers. Med Rec. 2022;4(2):196–202.
Tharwat M, Sakr NA, El-Sappagh S, Soliman H, Kwak K-S, Elmogy M. Colon cancer diagnosis based on machine learning and deep learning: modalities and analysis techniques. Sensors. 2022;22(23):9250. https://doi.org/10.3390/s22239250.
Li S, Yang Y, Wang X, et al. Colorectal cancer subtype identification from differential gene expression levels using minimalist deep learning. BioData Min. 2022;15:12. https://doi.org/10.1186/s13040-022-00295-w.
Srivastava A, Rai S, Singh M, Srivastava S. Computational intelligence-based gene expression analysis in colorectal cancer: a review. Comput Intell Oncol Stud Comput Intell. 2022. https://doi.org/10.1007/978-981-16-9221-5_22.
Al-Rajab M, Lu J, Xu Q. A framework model using multifilter feature selection to enhance colon cancer classification. PLoS ONE. 2021;16(4): e0249094.
Chan H, Chattopadhyay A, Chuang EY, Lu T. Development of a gene-based prediction model for recurrence of colorectal cancer using an ensemble learning algorithm. Front Oncol. 2021. https://doi.org/10.3389/fonc.2021.631056.
Rathore S, Hussain M, Ali A, Khan A. A recent survey on colon cancer detection techniques. IEEE/ACM Trans Comput Biol Bioinf. 2013;10(3):545–63. https://doi.org/10.1109/TCBB.2013.84.
Mulenga M, Kareem SA, Sabri AQM, Seera M. Stacking and chaining of normalization methods in deep learning-based classification of colorectal cancer using gut microbiome data. IEEE Access. 2021;9:97296–319.
Li H, Lin J, Xiao Y, Zheng W, Zhao L, Yang X, Zhong M, Liu H. Colorectal cancer detected by machine learning models using conventional laboratory test data. Technol Cancer Res Treatment. 2021. https://doi.org/10.1177/15330338211058352.
Kennion O, Maitland S, Brady R. Machine learning as a new horizon for colorectal cancer risk prediction? A systematic review. Health Sci Rev. 2022;4:100041.
Kinar Y, Kalkstein N, Akiva P, Levin B, Half EE, Goldshtein I, Chodick G, Shalev V. Development and validation of a predictive model for detection of colorectal cancer in primary care by analysis of complete blood counts: a binational retrospective study. J Am Med Inform Assoc. 2016;23(5):879–90.
Nartowt BJ, Hart GR, Muhammad W, Liang Y, Stark GF, Deng J. Robust machine learning for colorectal cancer risk prediction and stratification. Front Big Data. 2020. https://doi.org/10.3389/fdata.2020.00006.
Hoogendoorn M, Szolovits P, Moons LMG, Numans ME. Utilizing uncoded consultation notes from electronic medical records for predictive modeling of colorectal cancer. Artif Intell Med. 2016;69:53–61.
Kop R, Hoogendoorn M, ten Teije A, Büchner FL, Slottje P, Moons LMG, Numans ME. Predictive modeling of colorectal cancer using a dedicated pre-processing pipeline on routine electronic medical records. Comput Biol Med. 2016;76:30–8.
Nartowt BJ, Hart GR, Roffman DA, Llor X, Ali I, Muhammad W, Liang Y, Deng J. Scoring colorectal cancer risk with an artificial neural network based on self-reportable personal health data. PLoS ONE. 2019;14(8): e0221421.
Talukder MA, Islam MM, Uddin MA, Akhter A, Hasan KF, Moni MA. Machine learning-based lung and colon cancer detection using deep feature extraction and ensemble learning. Expert Syst Appl. 2022;205:117695.
Wang L. Predicting colorectal cancer using residual deep learning with nursing care. Contrast Media Mol Imaging. 2022;2022:7996195. https://doi.org/10.1155/2022/7996195.
Waljee AK, Weinheimer-Haus EM, Abubakar A, et al. Artificial intelligence and machine learning for early detection and diagnosis of colorectal cancer in sub-Saharan Africa. Gut. 2022;71:1259–65.
Paul S, Brahma D. An integrated approach for identification of functionally similar microRNAs in colorectal cancer. IEEE/ACM Trans Comput Biol Bioinf. 2019;16(1):183–92. https://doi.org/10.1109/TCBB.2017.2765332.
Ahn T, Kang N, Kim Y, Park T. Gene expression based prediction of prognostic outcome in ovarian cancer. In: 2018 IEEE international conference on bioinformatics and biomedicine (BIBM), Madrid, Spain, 2018, pp. 1753–1757.
Horaira MA, Ahmed MS, Kabir MH, Mollah MNH, Rahman Shah MA. Colon cancer prediction from gene expression profiles using kernel based support vector machine. In: 2018 International conference on computer, communication, chemical, material and electronic engineering (IC4ME2), Rajshahi, Bangladesh, 2018, pp. 1–4. https://doi.org/10.1109/IC4ME2.2018.8465636
Shafi ASM, Molla MMI, Jui JJ, et al. Detection of colon cancer based on microarray dataset using machine learning as a feature selection and classification techniques. SN Appl Sci. 2020;2:1243. https://doi.org/10.1007/s42452-020-3051-2.
Salem H, Attiya G, El-Fishawy N. Classification of human cancer diseases by gene expression profiles. Appl Soft Comput. 2017;50:124–34.
Nguyen T, Khosravi A, Creighton D, Nahavandi S. A novel aggregate gene selection method for microarray data classification. Pattern Recogn Lett. 2015;60–61:16–23.
Rani MJ, Devaraj D. Two-stage hybrid gene selection using mutual information and genetic algorithm for cancer data classification. J Med Syst. 2019. https://doi.org/10.1007/s10916-019-1372-8.
Acknowledgements
The authors acknowledged the Thanthai Periyar Govt. Arts & Science College (A), Tiruchirappalli, India for supporting the research work by providing the facilities.
Funding
No funding received for this research.
Author information
Authors and Affiliations
Contributions
This research endeavor was made possible by the collaboration and contributions of all authors.
Corresponding author
Ethics declarations
Conflict of Interest
No conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Butto, S.B., Bibi, K.F. Colon Cancer Detection Using Exhaustive Correlation Feature Selection Based Genetic Decision Support Regression. SN COMPUT. SCI. 6, 39 (2025). https://doi.org/10.1007/s42979-024-03561-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s42979-024-03561-2