Abstract
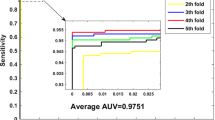
Protein–protein interactions (PPIs) are the basis to interpret biological mechanisms of life activity, and play vital roles in the execution of various cellular processes. The development of computer technology provides a new way for the effective prediction of PPIs and greatly arouses people’s interest. The challenge of this task is that PPIs data is typically represented in high-dimensional and is likely to contain noise, which will greatly affect the performance of the classifier. In this paper, we propose a novel feature weighted rotation forest algorithm (FWRF) to solve this problem. We calculate the weight of the feature by the \(\chi ^{2}\) statistical method and remove the low weight value features according to the selection rate. With this FWRF algorithm, the proposed method can eliminate the interference of useless information and make full use of the useful features to predict the interactions among proteins. In cross-validation experiment, our method obtained excellent prediction performance with the average accuracy, precision, sensitivity, MCC and AUC of 91.91, 92.51, 91.22, 83.84 and 91.60% on the H. pylori data set. We compared our method with other existing methods and the well-known classifiers, such as SVM and original rotation forest on the H. pylori data set. In addition, in order to demonstrate the ability of the FWRF algorithm, we also verified on the Yeast data set. The experimental results show that our method is more effective and robust in predicting PPIs. As a web server, the source code, H. pylori data sets and Yeast data sets used in this article are freely available at http://202.119.201.126:8888/FWRF/.







Similar content being viewed by others
References
Altschul SF, Madden TL, Schaffer AA et al (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25(17):3389–402
Bock JR, Gough DA (2003) Whole-proteome interaction mining. Bioinformatics 19(1):125–134
Enright AJ, Iliopoulos I, Kyrpides NC et al (1999) Protein interaction maps for complete genomes based on gene fusion events. Nature 402(6757):86–90
Gao ZG, Wang L, Xia SX et al (2016) Ens-PPI: a novel ensemble classifier for predicting the interactions of proteins using autocovariance transformation from PSSM. Biomed Res Int 2016(4):1–8
Gavin AC, Bosche M, Krause R et al (2002) Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415(6868):141–147
Gribskov M, McLachlan AD, Eisenberg D (1987) Profile analysis: detection of distantly related proteins. Proc Nat Acad Sci USA 84(13):4355–8
Guo Y, Yu L, Wen Z et al (2008) Using support vector machine combined with auto covariance to predict protein–protein interactions from protein sequences. Nucleic Acids Res 36(9):3025–3030
Ho Y, Gruhler A, Heilbut A et al (2002) Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415(6868):180–183
Ideker T, Ozier O, Schwikowski B et al (2002) Discovering regulatory and signalling circuits in molecular interaction networks. Bioinformatics (Oxford, England) 18 Suppl 1:S233-40
Ito T, Chiba T, Ozawa R et al (2001) A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc Nat Acad Sci USA 98(8):4569–4574
Ji Z, Wang B, Deng S et al (2014) Predicting dynamic deformation of retaining structure by LSSVR-based time series method. Neurocomputing 137:165–172
Jin Y (2000) Fuzzy modeling of high-dimensional systems: complexity reduction and interpretability improvement. IEEE Trans Fuzzy Syst 8(2):212–221
Jin Y, Sendhoff B (2008) Pareto-based multiobjective machine learning: an overview and case studies. IEEE Trans Syst Man Cybern Part C 38(3):397–415
Jin Y, Olhofer M, Sendhoff B (2002) A framework for evolutionary optimization with approximate fitness functions. IEEE Trans Evol Comput 6(5):481–494
Krogan NJ, Cagney G, Yu HY et al (2006) Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature 440(7084):637–643
Li Y, Olson EB (2010) A general purpose feature extractor for light detection and ranging data. Sensors 10(11):10356–10375
Li Y, Olson EB, IEEE (2011) Structure tensors for general purpose LIDAR feature extraction. In: IEEE international conference on robotics and automation ICRA, pp 1869–1874
Lin Z, You ZH, Huang DS et al (2013) t-LSE: a novel robust geometric approach for modeling protein-protein interaction networks. Plos One 8(4):e58368
Liu B, Yi J, Aishwarya SV et al (2013) QChIPat: a quantitative method to identify distinct binding patterns for two biological ChIP-seq samples in different experimental conditions. BMC Genom 14(8):S3
Mao Y, Xia Z, Yin Z et al (2007) Fault diagnosis based on fuzzy support vector machine with parameter tuning and feature selection. Chin J Chem Eng 15(2):233–239
Marcotte EM, Xenarios I, Eisenberg D (2001) Mining literature for protein–protein interactions. Bioinformatics 17(4):359–363
Martin S, Roe D, Faulon JL (2005) Predicting protein–protein interactions using signature products. Bioinformatics 21(2):218–226
Nanni L (2005) Hyperplanes for predicting protein–protein interactions. Neurocomputing 69(1–3):257–263
Nanni L, Lumini A (2006) An ensemble of K-local hyperplanes for predicting protein–protein interactions. Bioinformatics 22(10):1207–1210
Nanni L, Lumini A (2009) Ensemble generation and feature selection for the identification of students with learning disabilities. Expert Syst Appl 36(2):3896–3900
Ojansivu V, Heikkila J (2008) Blur insensitive texture classification using local phase quantization. Image Signal Process 5099:236–243
Pazos F, Valencia A (2001) Similarity of phylogenetic trees as indicator of protein–protein interaction. Protein Eng 14(9):609–614
Pazos F, Helmer-Citterich M, Ausiello G et al (1997) Correlated mutations contain information about protein–protein interaction. J Mol Biol 271(4):511–523
Rain JC, Selig L, De Reuse H et al (2001) The protein–protein interaction map of Helicobacter pylori (vol 409, pg 211, 2001). Nature 409(6821):743
Rodriguez JJ, Kuncheva LI (2006) Rotation forest: a new classifier ensemble method. IEEE Trans Pattern Anal Mach Intell 28(10):1619–1630
Shen J, Zhang J, Luo X et al (2007) Predictina protein–protein interactions based only on sequences information. Proc Nat Acad Sci USA 104(11):4337–4341
Theofilatos KA, Dimitrakopoulos CM, Tsakalidis AK et al (2011) Computational approaches for the prediction of protein–protein interactions: a survey. Curr Bioinform 6(4):398–414
Tuncbag N, Kar G, Keskin O et al (2009) A survey of available tools and web servers for analysis of protein–protein interactions and interfaces. Brief Bioinform 10(3):217–232
Wang H, Song A, Li B et al (2015) Psychophysiological classification and experiment study for spontaneous EEG based on two novel mental tasks. Technol Health Care 23:S249–S262
Xenarios I, Salwinski L, Duan XQJ et al (2002) DIP, the Database of Interacting Proteins: a research tool for studying cellular networks of protein interactions. Nucleic Acids Res 30(1):303–305
Yin Z, Zhou X, Bakal C et al (2008) Using iterative cluster merging with improved gap statistics to perform online phenotype discovery in the context of high-throughput RNAi screens. BMC Bioinform 9(1):264
Yin Z, Deng T, Peterson LE et al (2014) Transcriptome analysis of human adipocytes implicates the NOD-like receptor pathway in obesity-induced adipose inflammation. Mol Cell Endocrinol 394(1–2):80–87
You ZH (2010) Using manifold embedding for assessing and predicting protein interactions from high-throughput experimental data. Bioinformatics 26(21):2744–2751
You ZH, Yin Z, Han K et al (2010) A semi-supervised learning approach to predict synthetic genetic interactions by combining functional and topological properties of functional gene network. BMC Bioinform 11(1):343
You ZH, Lei YK, Zhu L et al (2013) Prediction of protein-protein interactions from amino acid sequences with ensemble extreme learning machines and principal component analysis. BMC Bioinform 14(8):1–11
You ZH, Zhou M, Luo X et al (2016) Highly efficient framework for predicting interactions between proteins. IEEE Trans Cyber 1–13
Zhang YQ, Zhang DL, Mi G et al (2012) Using ensemble methods to deal with imbalanced data in predicting protein–protein interactions. Comput Biol Chem 36:36–41
Zhu Z (2015) CompMap: a reference-based compression program to speed up read mapping to related reference sequences. Bioinformatics 31(3):426–8
Zhu H, Bilgin M, Bangham R et al (2001) Global analysis of protein activities using proteome chips. Science 293(5537):2101–2105
Zhu Z, Zhou J, Ji Z et al (2011) DNA sequence compression using adaptive particle swarm optimization-based memetic algorithm. IEEE Trans Evol Comput 15(5):643–658
Zhu Z, Zhang Y, Ji Z et al (2013a) High-throughput DNA sequence data compression. Brief Bioinform 16(1):1–15. doi:10.1093/bib/bbt087
Zhu L, You Z-H, Huang D-S (2013b) Increasing the reliability of protein-protein interaction networks via non-convex semantic embedding. Neurocomputing 121:99–107
Zhu Z, Jia S, He S et al (2015) Three-dimensional Gabor feature extraction for hyperspectral imagery classification using a memetic framework. Inf Sci 298(C):274–287
Zweig MH, Campbell G (1993) Receiver-operating characteristic (ROC) plots: a fundamental evaluation tool in clinical medicine. Clin Chem 39(4):561–77
Acknowledgements
This work is supported by the Fundamental Research Funds for the Central Universities (2017XKQY083).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there is no conflict of interests regarding the publication of this paper.
Additional information
Communicated by V. Loia.
Rights and permissions
About this article
Cite this article
Wang, L., You, ZH., Xia, SX. et al. An improved efficient rotation forest algorithm to predict the interactions among proteins. Soft Comput 22, 3373–3381 (2018). https://doi.org/10.1007/s00500-017-2582-y
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00500-017-2582-y