Abstract
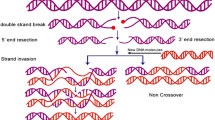
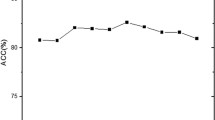
At the chromosomal level of evolution, recombination is a major factor for genetic variations. However, recombination does not occur with equal frequency at various regions of genome. The recombination has the tendency to occur at specific region with higher frequency and with low frequency at other regions, and former regions are named as hot recombination regions whereas later are called cold regions for recombination. In this paper, we have developed supervised machine learning-based models using artificial neural network, support vector machine and Naïve Bayes for efficient and effective classification of such hot and cold recombination regions based on the nucleotide composition of sequences. All models were validated and tested using tenfold cross-validation. Furthermore, neural network model was validated using leave one out and random sampling techniques in addition to tenfold cross-validation. Moreover, models were evaluated using receiver-operating curve. Our results indicate that artificial neural network achieves the best result.




Similar content being viewed by others
References
Hansen L, Kim N-K, Mariño-Ramírez L, Landsman D (2011) Analysis of biological features associated with meiotic recombination hot and cold spots in Saccharomyces cerevisiae. PLoS ONE 6(12):e29711
Smith GR (2001) Homologous recombination near and far from DNA breaks: alternative roles and contrasting views. Annu Rev Genet 35(1):243–274
Kauppi L, Jeffreys AJ, Keeney S (2004) Where the crossovers are: recombination distributions in mammals. Nat Rev Genet 5(6):413–424
Myers S, Bottolo L, Freeman C, McVean G, Donnelly P (2005) A fine-scale map of recombination rates and hotspots across the human genome. Science 310(5746):321–324
Baudat F, Nicolas A (1997) Clustering of meiotic double-strand breaks on yeast chromosome III. Proc Natl Acad Sci 94(10):5213–5218
Klein S, Zenvirth D, Dror V, Barton AB, Kaback DB, Simchen G (1996) Patterns of meiotic double-strand breakage on native and artificial yeast chromosomes. Chromosoma 105(5):276–284
Zenvirth D, Arbel T, Sherman A, Goldway M, Klein S, Simchen G (1992) Multiple sites for double-strand breaks in whole meiotic chromosomes of Saccharomyces cerevisiae. EMBO J 11(9):3441
Petes TD (2001) Meiotic recombination hot spots and cold spots. Nat Rev Genet 2(5):360–369
Kohl KP, Sekelsky J (2013) Meiotic and mitotic recombination in meiosis. Genetics 194(2):327–334
Lichten M, Goldman AS (1995) Meiotic recombination hotspots. Annu Rev Genet 29(1):423–444
Jeffreys AJ, Holloway JK, Kauppi L, May CA, Neumann R, Slingsby MT, Webb AJ (2004) Meiotic recombination hot spots and human DNA diversity. Philos Trans R Soc Lond B Biol Sci 359(1441):141–152
Wahls WP (1997) 2 Meiotic recombination hotspots: shaping the genome and Insights into hypervariable minisatellite DNA change. Curr Top Dev Biol 37:37–75
Gerton JL, DeRisi J, Shroff R, Lichten M, Brown PO, Petes TD (2000) Global mapping of meiotic recombination hotspots and coldspots in the yeast Saccharomyces cerevisiae. Proc Natl Acad Sci 97(21):11383–11390
Kliman RM, Irving N, Santiago M (2003) Selection conflicts, gene expression, and codon usage trends in yeast. J Mol Evol 57(1):98–109
Kliman RM, Hey J (1993) Reduced natural selection associated with low recombination in Drosophila melanogaster. Mol Biol Evol 10(6):1239–1258
Marais G, Mouchiroud D, Duret L (2001) Does recombination improve selection on codon usage? Lessons from nematode and fly complete genomes. Proc Natl Acad Sci 98(10):5688–5692
Marais G, Piganeau G (2002) Hill-Robertson interference is a minor determinant of variations in codon bias across Drosophila melanogaster and Caenorhabditis elegans genomes. Mol Biol Evol 19(9):1399–1406
Perry J, Ashworth A (1999) Evolutionary rate of a gene affected by chromosomal position. Curr Biol 9(17):987–989
Fullerton SM, Carvalho AB, Clark AG (2001) Local rates of recombination are positively correlated with GC content in the human genome. Mol Biol Evol 18(6):1139–1142
Friedel CC, Jahn KH, Sommer S, Rudd S, Mewes HW, Tetko IV (2005) Support vector machines for separation of mixed plant–pathogen EST collections based on codon usage. Bioinformatics 21(8):1383–1388
Bren U, Guengerich FP, Mavri J (2007) Guanine alkylation by the potent carcinogen aflatoxin B1: quantum chemical calculations. Chem Res Toxicol 20(8):1134–1140
Brown KL, Bren U, Stone MP, Guengerich FP (2009) Inherent stereospecificity in the reaction of aflatoxin B1 8, 9-epoxide with deoxyguanosine and efficiency of DNA catalysis. Chem Res Toxicol 22(5):913–917
Bren U, Fuchs JE, Oostenbrink C (2014) Cooperative binding of aflatoxin B1 by cytochrome P450 3A4: a computational study. Chem Res Toxicol 27(12):2136–2147
Biro JC (2008) Correlation between nucleotide composition and folding energy of coding sequences with special attention to wobble bases. Theor Biol Med Model 5(1):14
Bibb M, Findlay P, Johnson M (1984) The relationship between base composition and codon usage in bacterial genes and its use for the simple and reliable identification of protein-coding sequences. Gene 30(1):157–166
Ochman H, Lawrence JG, Groisman EA (2000) Lateral gene transfer and the nature of bacterial innovation. Nature 405(6784):299–304
Lin K, Kuang Y, Joseph JS, Kolatkar PR (2002) Conserved codon composition of ribosomal protein coding genes in Escherichia coli, Mycobacterium tuberculosis and Saccharomyces cerevisiae: lessons from supervised machine learning in functional genomics. Nucleic Acids Res 30(11):2599–2607
Liu G, Liu J, Cui X, Cai L (2012) Sequence-dependent prediction of recombination hotspots in Saccharomyces cerevisiae. J Theor Biol 293:49–54
Qiu W-R, Xiao X, Chou K-C (2014) iRSpot-TNCPseAAC: identify recombination spots with trinucleotide composition and pseudo amino acid components. Int J Mol Sci 15(2):1746–1766
Xia X, Xie Z (2001) DAMBE: software package for data analysis in molecular biology and evolution. J Hered 92(4):371–373
Carver T, Bleasby A (2003) The design of Jemboss: a graphical user interface to EMBOSS. Bioinformatics 19(14):1837–1843
Vapnik VN, Vapnik V (1998) Statistical learning theory, vol 2. Wiley, New York
Vapnik V (2000) The nature of statistical learning theory. Springer, Berlin
Burges CJ (1998) A tutorial on support vector machines for pattern recognition. Data Min Knowl Disc 2(2):121–167
Baxt WG, Shofer FS, Sites FD, Hollander JE (2002) A neural computational aid to the diagnosis of acute myocardial infarction. Ann Emerg Med 39(4):366–373
García-Pedrajas N, Hervás-Martínez C, Ortiz-Boyer D (2005) Cooperative coevolution of artificial neural network ensembles for pattern classification. IEEE Trans Evolut Comput 9(3):271–302
Yao X, Liu Y (1998) Making use of population information in evolutionary artificial neural networks. IEEE Trans Syst Man Cybern Part B Cybern 28(3):417–425
Demšar J, Curk T, Erjavec A, Gorup Č, Hočevar T, Milutinovič M, Možina M, Polajnar M, Toplak M, Starič A (2013) Orange: data mining toolbox in Python. J Mach Learn Res 14(1):2349–2353
Demšar J, Zupan B, Leban G, Curk T (2004) Orange: from experimental machine learning to interactive data mining. Springer, Berlin
Shafer G, Pearl J (1990) Readings in uncertain reasoning. Morgan Kaufmann Publishers Inc., California
Heckerman D, Geiger D, Chickering DM (1995) Learning Bayesian networks: the combination of knowledge and statistical data. Mach Learn 20(3):197–243
Jensen FV (1996) An introduction to Bayesian networks, vol 210. UCL press, London
Peral J (1988) Probabilistic reasoning in intelligent systems, vol 12. Morgan Kaufmann, California, pp 241–288
Castillo E (1997) Expert systems and probabilistic network models. Springer, Berlin
Metz CE (1978) Basic principles of ROC analysis. In: Seminars in nuclear medicine, vol 4. Elsevier, pp 283–298
Kanmani S, Uthariaraj VR, Sankaranarayanan V, Thambidurai P (2007) Object-oriented software fault prediction using neural networks. Inf Softw Technol 49(5):483–492
Briand LC, Wüst J (2002) Empirical studies of quality models in object-oriented systems. Adv Comput 56:97–166
Acknowledgments
The authors are highly grateful to Department of Biotechnology, New Delhi for providing support for this work under Bioinformatics Infrastructure Facility of DBT at MANIT Bhopal.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Dwivedi, A.K., Chouhan, U. Comparative study of artificial neural network for classification of hot and cold recombination regions in Saccharomyces cerevisiae . Neural Comput & Applic 29, 529–535 (2018). https://doi.org/10.1007/s00521-016-2466-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00521-016-2466-6