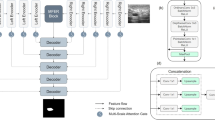
Abstract
Medical image segmentation plays an important role in many clinical medicines, such as medical diagnosis and computer-assisted treatment. However, due to the large quality differences, variable lesion areas and their complex shapes, medical image segmentation is a very challenging task. However, most of the recent deep learning methods ignore the global context information as well as the receptive fields of pixels and do not consider the reuse of pixel features during the feature extraction stage. In this paper, we propose DGFAU-Net, an encoder–decoder structured 2D segmentation model, to overcome the shortcomings aforementioned. In the encoder, DenseNet and AtrousCNN networks are leveraged to extract image features. The DenseNet network is mainly used to achieve the reuse of pixel features, and AtrousCNN is utilized to enhance the receptive field of pixels. In the decoder, two modules, global feature attention upsample (GFAU) and pyramid pooling attention squeeze-excitation (PPASE), are proposed. GFAU combines low-level and high-level features to generate new features with richer information for improving the perceptions of global contextual information of pixels. PPASE employs a multi-scale pooling layer to enhance the pixel’s acceptance field. In addition, Focal loss is used to balance the difference between samples of the lesion and non-lesioned areas. Extensive experiments are conducted on one local dataset and two public datasets, which are the local dataset of MRI images of carotid plaque, DRIVE vessel segmentation dataset and CHASE_DB1 vessel segmentation dataset, and the experimental results demonstrate the effectiveness of our proposed model.







Similar content being viewed by others
References
Huang G, Liu Z, Weinberger KQ (2016) Densely Connected Convolutional Networks, CoRR abs/1608.06993
Noh H, Hong S, Han B Learning Deconvolution Network for Semantic Segmentation, in ICCV (IEEE Computer Society, 2015), pp. 1520–1528
Badrinarayanan V, Kendall A, Cipolla R (2017) Deep Convolutional Encoder-Decoder Architecture for Image Segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 39(12):2481
An F, Liu Z (2019) Medical image segmentation algorithm based on feedback mechanism convolutional neural network’ Biomed. Signal Process Control. https://doi.org/10.1016/j.bspc.2019.101589
Calisto MGB, Lai-Yuen SK (2020) AdaResU-Net: Multiobjective adaptive convolutional neural network for medical image segmentation. Neurocomputing 392:325. https://doi.org/10.1016/j.neucom.2019.01.110
Chen L, Papandreou G, Kokkinos I, Murphy K, Yuille AL (2015) Semantic Image Segmentation with Deep Convolutional Nets and Fully Connected CRFs ,in ICLR (Poster)
Ronneberger O, Fischer P (2015) Brox T in MICCAI (3). Lect Notes Comput Sci 9351:234–241
Drozdzal M, Vorontsov E, Chartrand G, Kadoury S, Pal C (2016) in LABELS/DLMIA@MICCAI, Lecture Notes in Computer Science, vol. 10008. Lect Notes Comput Sci 10008:179–187
Lin G, Milan A, Shen C, Reid I.D RefineNet: Multi-path Refinement Networks for High-Resolution Semantic Segmentation, in CVPR (IEEE Computer Society, 2017), pp. 5168–5177
Peng C, Zhang X, Yu G, Luo G, Sun J Large Kernel Matters - Improve Semantic Segmentation by Global Convolutional Network, in CVPR (IEEE Computer Society, 2017), pp. 1743–1751
Chen L, Papandreou G, Schroff F, Adam H Rethinking Atrous Convolution for Semantic Image Segmentation, CoRR abs/1706.05587 (2017)
Chen L, Papandreou G, Kokkinos I, Murphy K, Yuille AL (2018) DeepLab: Semantic Image Segmentation with Deep Convolutional Nets, Atrous Convolution, and Fully Connected CRFs. IEEE Trans Pattern Anal Mach Intell 40(4):834. https://doi.org/10.1109/TPAMI.2017.2699184
Zhao H, Shi J, Qi X, Wang X, Jia J Pyramid Scene Parsing Network, in CVPR (IEEE Computer Society, 2017), pp. 6230–6239
Hu J, Shen L, Sun G Squeeze-and-Excitation Networks, in CVPR (IEEE Computer Society, 2018), pp. 7132–7141
Yu F, Koltun V (2016) in ICLR (Poster)
Soares JVB, Leandro JJG, Cesar RM, Jelinek HF, Cree MJ (2006) Retinal vessel segmentation using the 2-D Gabor wavelet and supervised classification. IEEE Trans Med Imaging 25(9):1214
Azzopardi G, Strisciuglio N, Vento M, Petkov N (2015) Trainable COSFIRE filters for vessel delineation with application to retinal images. Med Image Anal 19(1):46
Li Q, Feng B, Xie L, Liang P, Zhang H, Wang T (2016) A Cross-Modality Learning Approach for Vessel Segmentation in Retinal Images. IEEE Trans. Med. Imaging 35(1):109
Tian Z, Liu L, Zhang Z, Fei B (2016) Superpixel-Based Segmentation for 3D Prostate MR Images. IEEE Trans Med Imaging 35(3):791
Fang L, Wang X, Wang L (2020) Multi-modal medical image segmentation based on vector-valued active contour models. Inf Sci 513:504. https://doi.org/10.1016/j.ins.2019.10.051
Miao J, Zhou X, Huang T (2020) Local segmentation of images using an improved fuzzy C-means clustering algorithm based on self-adaptive dictionary learning. Appl Soft Comput. https://doi.org/10.1016/j.asoc.2020.106200
Pandey S, Singh PR, Tian J (2020) An image augmentation approach using two-stage generative adversarial network for nuclei image segmentation. Biomed Signal Process Control. https://doi.org/10.1016/j.bspc.2019.101782
Göçeri E, Shah ZK, Layman RR, Jiang X, Gurcan MN (2016) Quantification of liver fat: A comprehensive review. Comp in Bio Med 71:174
Gao K, Niu S, Ji Z, Wu M, Chen Q, Xu R, Yuan S, Fan W, Chen Y, Dong J (2019) Double-branched and area-constraint fully convolutional networks for automated serous retinal detachment segmentation in SD-OCT images. Comput Methods Programs Biomed 176:69. https://doi.org/10.1016/j.cmpb.2019.04.027
Long J, Shelhamer E, Darrell T in CVPR (IEEE Computer Society, 2015), pp. 3431–3440
Karthik R, Gupta U, Jha A, Rajalakshmi R, Menaka R (2019). Appl Soft Comput. https://doi.org/10.1016/j.asoc.2019.105685
Hosseini B, Montagné R, Hammer B (2020) Deep-Aligned Convolutional Neural Network for Skeleton-Based Action Recognition and Segmentation. Data Sci Eng 5(2):126. https://doi.org/10.1007/s41019-020-00123-3
Jiang F, Grigorev A, Rho S, Tian Z, Fu Y, Sori WJ, Khan A, Liu S (2018) Medical image semantic segmentation based on deep learning. Neural Comput Appl 29(5):1257. https://doi.org/10.1007/s00521-017-3158-6
Fraz MM, Khurram SA, Graham S, Shaban M, Hassan M, Loya A, Rajpoot NM (2020) FABnet: feature attention-based network for simultaneous segmentation of microvessels and nerves in routine histology images of oral cancer. Neural Comput Appl 32(14):9915. https://doi.org/10.1007/s00521-019-04516-y
Ji Y, Zhang H, Jie Z, Ma L, Wu QMJ (2020). IEEE Transactions on Neural Networks and Learning Systems. https://doi.org/10.1109/TNNLS.2020.3007534
Tang W, Zou D, Yang S, Shi J, Dan J, Song G (2020) A two-stage approach for automatic liver segmentation with Faster R-CNN and DeepLab. Neural Comput Appl 32(11):6769. https://doi.org/10.1007/s00521-019-04700-0
Fu H, Xu Y, Lin S, Wong DWK, Liu J (2016) in Medical Image Computing and Computer-Assisted Intervention - MICCAI 2016–19th International Conference, Athens, Greece, October 17–21 Proceedings. Part II 2016:132–139. https://doi.org/10.1007/978-3-319-46723-8_16
Norman B, Pedoia V, Noworolski A, Link TM, Majumdar S (2019) Applying Densely Connected Convolutional Neural Networks for Staging Osteoarthritis Severity from Plain Radiographs. J. Digital Imaging 32(3):471. https://doi.org/10.1007/s10278-018-0098-3
Li H, Xiong P, An J, Wang L (2018) BMVC. BMVA Press, p 285
Huang Z, Wang X, Huang L, Huang C, Wei Y, Liu W (2018) CoRR abs/1811.11721 . http://arxiv.org/abs/1811.11721
Zhang C, Zhu L, Zhang S (2019) CoRR abs/1906.01792 . http://arxiv.org/abs/1906.01792
Wang L, Yu X, Bourlai T, Metaxas DN (2019) A coupled encoder-decoder network for joint face detection and landmark localization. Image Vis Comput 87:37. https://doi.org/10.1016/j.imavis.2018.09.008
Kim Y, Kim KR, Choe YH (2020) Automatic myocardial segmentation in dynamic contrast enhanced perfusion MRI using Monte Carlo dropout in an encoder-decoder convolutional neural network. Comput. Methods Programs Biomed. 185. https://doi.org/10.1016/j.cmpb.2019.105150
Andreini P, Bonechi S, Bianchini M, Mecocci A, Scarselli F (2020) Image generation by GAN and style transfer for agar plate image segmentation. Comput Methods Programs Biomed. https://doi.org/10.1016/j.cmpb.2019.105268
Liu H, Fu Z, Han J, Shao L, Hou S, Chu Y (2019) Single image super-resolution using multi-scale deep encoder-decoder with phase congruency edge map guidance. Inf Sci 473:44. https://doi.org/10.1016/j.ins.2018.09.018
Sun Y, Tian Y, Xu Y (2019) Problems of encoder-decoder frameworks for high-resolution remote sensing image segmentation: Structural stereotype and insufficient learning. Neuro comput 330:297. https://doi.org/10.1016/j.neucom.2018.11.051
He K (2016) Zhang X. Sun J in CVPR (IEEE Computer Society, Ren S, pp 770–778
Kaiming He XZ, Ren S (2016) Sun J in ECCV (4). Lect Notes Comput Sci 9908:630–645
Ioffe S, Szegedy C in ICML, JMLR Workshop and Conference Proceedings, vol. 37 (JMLR.org, 2015), JMLR Workshop and Conference Proceedings, vol. 37, pp. 448–456
Yang M, Yu K (2018) Zhang C. Yang K in CVPR (IEEE Computer Society, Li Z, pp 3684–3692
Staal J, Abràmoff MD, Niemeijer M, Viergever MA, van Ginneken B (2004) Ridge-based vessel segmentation in color images of the retina. IEEE Trans Med Imaging 23(4):501
CHASE\_DB1 dataset. https://blogs.kingston.ac.uk/retinal/chasedb1/,
Roychowdhury S, Koozekanani D, Parhi KK (2015) Blood Vessel Segmentation of Fundus Images by Major Vessel Extraction and Subimage Classification. IEEE J Biomed Health Inform 19(3):1118
Ni J, Wu J, Tong J, Chen Z, Zhao J (2019) Computer Methods and Programs in Biomedicine
Acknowledgements
The work is supported by the National Natural Science Foundation of China under Grant No. 61772342. We would like to express our special thanks to the members in our laboratory for their valuable advice on this work.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
No conflict of interest needs to be declared.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Funding
This study was funded by the National Natural Science Foundation of China (grant number 61772342).
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Peng, D., Yu, X., Peng, W. et al. DGFAU-Net: Global feature attention upsampling network for medical image segmentation. Neural Comput & Applic 33, 12023–12037 (2021). https://doi.org/10.1007/s00521-021-05908-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00521-021-05908-9