Abstract
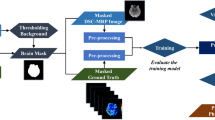
Susceptibility-weighted images (SWIs) have recently been confirmed to be more sensitive to acute ischemic stroke than diffusion weighted images in the form of the presence of small veins shortly after the symptom onset. Accurate segmentation of small veins in SWIs is critical for quantitative diagnosis, individual therapy and outcome prediction of acute ischemic stroke. It is challenging to segment veins in SWIs as they exhibit substantial variability and intensity inhomogeneity within a patient and among patients, which may even be hard for experts to delineate manually. A deep convolutional neural network is proposed to segment veins in SWIs with two main contributions: dense connection to concatenate feature maps from preceding layers to enhance network performance, and a hybrid loss function comprising of classification accuracy and global region overlap terms to handle class imbalance. Experiments have been conducted on 10 consecutive patients with acute ischemic stroke using leave-one-out validation, yielding the best Dice coefficient (0.756 ± 0.043) (p < 0.001) as compared with 3 relevant methods. The proposed method could provide a potential tool to quantify veins in SWIs with accuracy to assist decision making especially for thrombolytic therapy.






Similar content being viewed by others
References
Lin MP, Liebeskind DS (2016) Imaging of ischemic stroke. Continuum 22(5):1399
Niu SZ, Yu GH, Ma JH, Wang J (2018) Nonlocal low-rank and sparse matrix decomposition for spectral CT reconstruction. Inverse Probl 34(2):024003
Niu SZ, Huang J, Bian ZY, Zeng D, Chen WF, Yu GH, Liang ZR, Ma JH (2017) Iterative reconstruction for sparse-view x-ray CT using alpha-divergence constrained total generalized variation minimization. J X-ray Sci Technol 25(4):673–688
Niu SZ, Gao Y, Bian ZY, Huang J, Chen WF, Yu GH, Liang ZR, Ma JH (2014) Sparse-view x-ray CT reconstruction via total generalized variation regularization. Phys Med Biol 59(12):2997
Niu SZ, Zhang SL, Huang J, Bian ZY, Chen WF, Yu GH, Liang ZR, Ma JH (2016) Low-dose cerebral perfusion computed tomography image restoration via low-rank and total variation regularizations. Neurocomputing 197:143–160
Chalian M, Tekes A, Meoded A, Poretti A, Huisman TA (2011) Susceptibility-weighted imaging (SWI): a potential non-invasive imaging tool for characterizing ischemic brain injury? J Neuroradiol 38:187–190
Kao HW, Tsai FY, Hasso AN (2012) Predicting stroke evolution: comparison of susceptibility-weighted MR imaging with MR perfusion. Eur Radiol 22(7):1397–1403
Heyn C, Alcaide-Leon P, Bharatha A, Sussman MS, Kucharczyk W, Mandell DM (2016) Susceptibility-weighted imaging in neurovascular disease. Top Magn Reson Imaging 25(2):63–71
Chen CY, Chen CI, Tsai FY, Tsai PH, Chan WP (2015) Prominent vessel sign on susceptibility-weighted imaging in acute stroke: Prediction of infarct growth and clinical outcome. PLoS ONE 10(6):e0131118
Baik SK, Choi W, Oh SJ, Park KP, Park MG, Yang TI, Jeong HW (2012) Change in cortical vessel signs on susceptibility-weighted images after full recanalization in hyperacute ischemic stroke. Cerebrovasc Dis 34:206–212
Sun W, Liu W, Zhang Z, Xiao L, Duan Z, Liu D, Xiong Y, Zhu W, Lu G, Liu X (2014) Asymmetrical cortical vessel sign on susceptibility-weighted imaging: a novel imaging marker for early neurological deterioration and unfavorable prognosis. Eur J Neurol 21:1411–1418
Zhao GJ, Sun L, Wang ZR, Wang LQ, Cheng ZR, Lei HR, Yang DQ, Cui YS, Zhang SR (2017) Evaluation of the role of susceptibility-weighted imaging in thrombolytic therapy for acute ischemic stroke. J Clin Neurosci 40:175–179
Lou M, Chen Z, Wan J, Hu H, Cai X, Shi Z, Sun J (2014) Susceptibility-diffusion mismatch predicts thrombolytic outcomes: a retrospective cohort study. Am J Neuroradiol 35:2061–2067
Haacke EM, Xu YB, Cheng YC, Reichenbach JR (2004) Susceptibility weighted imaging (SWI). Magn Reson Med 52:612–618
Lesage D, Angelini ED, Bloch I, Funka-Lea G (2009) A review of 3D vessel lumen segmentation techniques: models, features and extraction schemes. Med Image Anal 13(6):819–845
Hassouna MS, Farag AA, Hushek S, Moriarty T (2006) Cerebrovascular segmentation from TOF using stochastic models. Med Image Anal 10(1):2–18
Babin D, Pižurica A, De Vylder J, Vansteenkiste E, Philips W (2013) Brain blood vessel segmentation using line-shaped profiles. Phys Med Biol 58(22):8041–8061
Frangi AF, Niessen WJ, Vincken KL, Viergever MA (1998) Multiscale vessel enhancement filtering. MICCAI, LNCS 1496:130–137
Shang YF, Deklerck R, Nyssen E, Markova A, de Mey J, Yang X, Sun K (2011) Vascular active contour for vessel tree segmentation. IEEE Trans Biomed Eng 58(4):1023–1032
Bogunović H, Pozo JM, Villa-Uriol MC, Majoie CB, van den Berg R, Gratama van Andel HA, Macho JM, Blasco J, Román LS, Frangi AF (2011) Automated segmentation of cerebral vasculature with aneurysms in 3DRA and TOF-MRA using geodesic active regions: an evaluation study. Med Phys 38(1):210–222
Bériault S, Archambault-Wallenburg M, Sadikot AF, Collins DL, Pike GB (2014) Automatic markov random field segmentation of susceptibility-weighted MR venography. LNCS 8361:39–47
Bériault S, Xiao YM, Collins DL, Pike GB (2015) Automatic SWI venography segmentation using conditional random fields. IEEE Trans Med Imaging 34(12):2478–2491
Litjens G, Kooi T, Bejnordi BE, Setio AAA, Ciompi F, Ghafoorian M, van derLaak JAWM, van Ginneken B, Sánchez CI (2017) A survey on deep learning in medical image analysis. Med Image Anal 42:60–88
LeCun Y, Boser B, Denker JS, Henderson D, Howard RE, Hubbard W, Jackel LD (1989) Backpropagation applied to handwritten zip code recognition. Neural Comput 1(4):541–551
Krizhevsky A, Sutskever I, Hinton GE (2012) Imagenet classification with deep convolutional neural networks. In: International conference on neural information processing systems, pp 1097–1105
Ciresan D, Giusti A, Gambardella LM, Schmidhuber J (2012) Deep neural networks segment neuronal membranes in electron microscopy images. In: International conference on neural information processing systems, pp 2843–2851
Long J, Shelhamer E, Darrell T (2015) Fully convolutional networks for semantic segmentation. In: IEEE conference on computer vision and pattern recognition, pp 3431–3440
Ronneberger O, Fischer P, Brox T (2015) U-net: convolutional networks for biomedical image segmentation. In: International conference on medical image computing and computer-assisted intervention, pp 234–241
Çiçek Ö, Abdulkadir A, Lienkamp SS, Brox T, Ronneberger O (2016) 3D U-Net: learning dense volumetric segmentation from sparse annotation. In: International conference on medical image computing and computer-assisted intervention, pp 424–32
Milletari F, Navab N, Ahmadi SA (2016) V-net: fully convolutional neural networks for volumetric medical image segmentation. In: IEEE international conference on 3D vision (3DV), pp 565–571
Chen H, Dou Q, Yu LQ, Qin J, Heng PA (2017) VoxResNet: deep voxelwise residual networks for brain segmentation from 3D MR images. NeuroImage 170:446–455
He KM, Zhang XY, Ren SQ, Sun J (2016) Deep residual learning for image recognition. In: IEEE conference on computer vision and pattern recognition, pp 770–778
He KM, Zhang XY, Ren SQ, Sun J (2016) Identity mappings in deep residual networks. In: European conference on computer vision 2016, pp 630–645
Kamnitsas K, Ledig C, Newcombe VFJ, Simpson JP, Kane AD, Menon DK, Rueckert D, Glocker B (2016) Efficient multi-scale 3D CNN with fully connected CRF for accurate brain lesion segmentation. Med Image Anal 36:61–78
Kamnitsas K, Ferrante E, Parisot S, Ledig C, Nori A, Criminisi A, Rueckert D, Glocker B (2016) DeepMedic for brain tumor segmentation. In: International workshop on Brainlesion: Glioma, multiple sclerosis, stroke and traumatic brain injuries, pp 138–149
Huang G, Liu Z, van der Maaten L, Weinberger KQ (2017) Densely connected convolutional networks. IEEE Conf Comput Vis Pattern Recogn 1(2):3
Simonyan K, Zisserman A (2014) Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:1409.1556
Ioffe S, Szegedy C (2015) Batch normalization: accelerating deep network training by reducing internal covariate shift. In: International conference on machine learning, pp 448–456
He KM, Zhang XY, Ren SQ, Sun J (2015) Delving deep into rectifiers: Surpassing human-level performance on imagenet classification. In: IEEE international conference on computer vision, pp 1026–1034
Wolf I, Vetter M, Wegner I, Böttger T, Nolden M, Schöbinger M, Hastenteufel M, Kunert T, Meinzer HP (2005) The medical imaging interaction toolkit. Med Image Anal 9(6):594–604
Sutskever I, Martens J, Dahl G, Hinton G (2013) On the importance of initialization and momentum in deep learning. In: International conference on machine learning, pp 1139–1147
Srivastava N, Hinton G, Krizhevsky A, Sutskever I, Salakhutdinov R (2014) Dropout: a simple way to prevent neural networks from overfitting. J Mach Learn Res 15(1):1929–1958
Maier O, Menze BH, von der Gablentz J et al (2017) ISLES 2015-A public evaluation benchmark for ischemic stroke lesion segmentation from multispectral MRI. Med Image Anal 35:250–269
Acknowledgements
This work has been supported by: National Key R&D Program of China 2018YFB1105600, National Natural Science Foundation of China 61671440, Shenzhen Peacock Technology Innovation Project KQJSCX20170731162830878, National Natural Science Foundation of China 61672510, and Shenzhen Science and Technology Program JCYJ20160331191401141. The authors wish to thank Dr. Ahmed Elazab from Shenzhen University for his help in enhancing the language usage.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
We declare that all authors have no conflicts of interest in the authorship or publication of this contribution.
Rights and permissions
About this article
Cite this article
Zhang, X., Zhang, Y. & Hu, Q. Deep learning based vein segmentation from susceptibility-weighted images. Computing 101, 637–652 (2019). https://doi.org/10.1007/s00607-018-0677-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00607-018-0677-7
Keywords
- Ischemic stroke
- Susceptibility-weighted image
- Convolutional neural network
- Dense connection
- Hybrid loss function