Abstract
In this paper we present a novel methodology based on non-parametric deformable prototype templates for reconstructing the outline of a shape from a degraded image. Our method is versatile and fast and has the potential to provide an automatic procedure for classifying pathologies. We test our approach on synthetic and real data from a variety of medical and biological applications. In these studies it is important to reconstruct accurately the shape of the object under investigation from very noisy data. Here we assume that we have some prior knowledge about the object outline represented by a prototype shape. Our procedure deforms this shape by means of non-affine transformations and the contour is reconstructed by minimizing a newly developed objective function that depends on the transformation parameters. We introduce an iterative template deformation procedure in which the scale of the deformation decreases as the algorithm proceeds. We compare our results with those from a Gaussian Mixture Model segmentation and two state-of-the-art Level Set methods. This comparison shows that the proposed procedure performs consistently well on both real and simulated data. As a by-product we develop a new filter that recovers the connectivity of a shape.








Similar content being viewed by others
Notes
The SNR in decibels is given by SNR = 20 log10(V s/V n), where V s is the signal strength and V n is the noise level. In this study we considered the difference \(\mu_{\texttt{I}}-\mu_{\texttt{E}}\) between the means of A inside and outside the shape as signal strength V s and the standard deviation of the noise distribution as noise level V n.
References
McInerney T, Terzopoulos D (1996) Deformable models in medical image analysis: a survey. Med Image Anal 1(2):91–108
Baradez MO, McGuckin CP, Forraz N, Pettengel R, Hoppe A (2004) Robust and automed unimodal histogram thresholding and potential applications. Pattern Recognit 37:1131–1148
Qu YD, Cui CS, Chen SB, Li JQ (2005) A fast subpixel edge detection method using sobel-zernike moments operator. Image Vis Comput 23:11–17
Hamid MR, Baloch A, Bilal A, Zaffar N (2003) Object segmentation using feature based conditional morphology. In: Proceedings of the 12th international conference on IAP, pp 548–553
Chung Kl, Huang HL, Lu HI (2004) Efficient region segmentation on compressed gray images using quadtree and shading representation. Pattern Recognit 37:1591–1605
Gunsel B, Jain AK, Panayirci E (1996) Reconstruction and boundary detection of range and intensity images using multiscale MRF representations. IEEE Trans Med Imaging 63(2):353–366
Antoine JP, Barache D, Cesar RM Jr, da Fontoura Costa L (1997) Shape characterization with the wavelet transform. Signal Process 62:265–290
Wong HS, Caelli T, Guan L (2000) A model-based Neural Network for Edge Characterization. Pattern Recognit 33:427–444
Ho SY, Lee KZ (2003) Design and analysis of an efficient evolutionary image segmentation algorithm. J VLSI Signal Process 35:29–42
Jain AK, Zhong Y, Jolly MPD (1998) Deformable template models: a review. Signal Process 71(22):109–129
Amit Y, Manbeck KM (1993) Deformable template models for emission tomography. IEEE Trans Med Imaging 12(2):260–268
Zagrosdsy V, Walimbe V, Castro-Pareja CR, Qin JX, Son JM, Shekhar R (2005) Registration assisted segmentation of real time 3D echocardiographic data using deformable models. IEEE Trans Med Imaging 24(9):1089–1099
Kass M, Witkin A, and Terzopoulos D (1988) Snakes: active contour models. Int J Comput Vis 1:321–331
Terzopolous D, Witkin A, and Kass M (1988) Contraints on deformable models: recovering 3D shape and non-rigid motion. Artif Intell 36(1):91–123
Cohen LD, Cohen I (1993) Finite-element methods for active contour and balloons for 2D and 3D images. IEEE Trans Patt Anal Mach Intell 15:131–147
Lemarie F, Levine M (1993) Tracking deformable objects in the plane using active contour model. IEEE Trans Patt Anal and Mach Intell 15(6):617–634
Yuille AL, Hallinan PW, Cohen DS (1992) Feature extraction from faces using deformable templates. Int J Comput Vis 8(2):133–144
Kindratenko VV (2003) On using functions to describe the shape. J Math Image Vis 18:225–245
Grenander U, Keenan DM (1993) Advances in applied statistics: statistics and images. Carfax Publishing Company, Abingdon
Hurn M (1998) Confocal fluorescence microscopy of leaf cells: an application of Bayesian image analysis. Appl Stat 47:361–377
Baumberg A, Hogg D (1995) An adaptive eigenshape model. In: Proceedings of the 6th British machine vision conference, vol 15, pp 617–634
Haddania J, Faez K, Moallem P (2001) Neural network based face recognition with moment invariants. In: International conference on image processing
Amit A, Grenander U, Piccioni M (1991) Structural image restoration through deformable templates. J Am Stat Assoc 86(414):376–388
Jain AK, Zhong Y, Lakshmanan S (1996) Object matching using deformable templates. IEEE Trans Patt Anal Mach Intell 18(3):267–277
de Pasquale F, Barone P, Sebastiani G, Stander J (2004) Bayesian analysis of dynamic magnetic resonance breast images. Appl Stat 53(3):475–493
Heywang-Kobrunner SH, Beck R (1995) Contrast enhanced MRI of the breast. Springer, Berlin
Hobolth A, Jensen EBV (2000) Modelling stochastic changes in curve shape, with an application to cancer diagnostics. Adv Appl Probab 32:344–362
Blekas K, Likas A, Galatsanos N, Lagaris I (2005) A spatially constrained mixture model for image segmentation. IEEE Trans Neural Net 16(2):494–498
Debreuve ‘E, Gastaud M, Barlaud M, Aubert G (2007) Using the shape gradient for active contour segmentation: from the continuous to the discrete formulation. J Math Imaging Vis 28:47–66
Li C, Xu C, Gui C, Fox M (2005) Level set evolution without re-inizialitation: a new variational formula. In: Proceedings of CVPR05
Parker JR (1997) Algorithms for image processing and computer vision. Wiley, New York
Hu MK (1962) Visual pattern recognition by moment invariants. IEEE Trans Inform Theory 8:179–187
Butkov E (1968) Mathematical physics. AW, Reading
Hoel PG, Port SC, and Stone CJ (1971) Introduction to statistical theory. Houghton Mifflin, New York
Soille P (1999) Morphological image analysis. Springer, Berlin
Acknowledgments
We thank Drs. Giovanni Sebastiani and Piero Barone for their considerable contribution and helpful advice. We are grateful to radiologists at the Santa Lucia Hospital, Rome, Italy for their valuable collaboration. The Biomedical Ultrasonics Laboratory, Biomedical Engineering Department, University of Michigan, USA provided the Ultrasound data, while Dr. Li, Department of Electrical and Computer Engineering, University of Connecticut Storrs, CT, USA, provided the cell and US data. Dr. Barre’ provided the NM heart images. We acknowledge the Department of Radiology at Brigham and Women’s Hospital, Boston, USA for providing the CT image. Comments from the Associate Editor and the Reviewers have considerably enhanced and improved this paper.
Author information
Authors and Affiliations
Corresponding author
Appendix
Appendix
1.1 A1. Range of deformation parameters
For a detailed derivation of the deformation model, see [33]. Here we start from:
in which \({\varvec{\varepsilon}}=\left[\varepsilon^{1},\varepsilon^{2}\right],\; {\varvec{\psi}}=\left[2 \sin \left( \pi x \right) \cos \left( \pi y \right),2 \cos \left( \pi x \right) \sin \left( \pi y \right) \right]\) and \({\varvec{ \Uptheta}}=(\alpha;{\varvec{\varepsilon}}).\) In order to widen the class of deformations that we consider, we shall assume that ɛ 1 nm and ɛ 2 nm can take any value in [−1, 1] independently of each other.
We now discuss suitable ranges for the deformation parameters. Since we want every point of S to be mapped within the unit square, the minimum value of α can be obtained by requiring that the maximum deformation in both directions will never leave the unit square. We discuss this for the x direction; an analogous treatment applies for the y direction. First of all, we consider a point (x, y) very close to the border (0, y). We require that this point will be mapped within S, so that applying the maximum negative deformation in the x direction must yield a non-negative transformed point. The maximum negative deformation corresponding to ɛ1 = −1 must satisfy:
Since x is small we approximate sin(πx) by πx, and in order to have the biggest deformation we set cos(πy) = 1. With these simplifications (8) becomes:
from which it follows that
Analogously, if we take a point close to the border (1, y) we require that the biggest positive deformation corresponding to ɛ1 = 1 will not map this point beyond the border so that:
Now, since x is very close to 1, we can approximate sin(π x) by π(1 − x) and as before in order to have the biggest deformation we take cos(πy) = 1. Hence, (9) becomes:
leading again to the same constraint
Now, because of the discretization of the mapping we will assume the value 0.4 for the minimum of α. The maximum value of α can be obtained by requiring that the maximum deformation is greater than the distance between two adjacent pixels. This happens when
because the pixel grids that we use are typically of size 50 × 50. In this way, if
every point of S will be mapped into itself and no effect of the deformation will be visible. In conclusion, we adopted the range [0.4, 5] for the scale parameter α.
1.2 A2. The power of the test
We choose w by considering the power of the test:
In particular, let us require power λ when
That is, we require P(W ≥ w | W ∼ N(δ, 1)) = λ. Hence, P(W − δ ≥ w − δ| W − δ ∼ N(0, 1)) = λ, so that 1 − Φ(w − δ) = λ, or w = δ + Φ−1(1 − λ), where Φ is the cumulative distribution function of a standard normal variable.
An upper bound on the probability of Type I error can be found as follows:
assuming that under H 0 the probability that both pixels belong to I (or E ) is \({\frac{1}{2}}.\) Elementary calculus leads us to the bound:
with equality holding when \(\sigma_{\texttt{I}}=\sigma_{\texttt{E}}.\) Hence, an upper bound for the probability of Type I error is 1 − Φ{δ + Φ−1(1 − λ ) }. In our simulation study a conservative value of δ is 1.5. Thus, for power λ = 0.7, we can set w = 1. This choice leads to an upper bound for the probability of Type I error of 0.16, which is acceptable.
1.3 A3. A new filter to recover the connectivity
Although the operator \({{\mathcal{K}}}\) in (1) preserves the connectivity of the template on a continuous space, it may not always do so on a discrete space such as the space of image pixels. Because of the definition of the objective function given in Sect. 2.3, to perform our analysis we need a simply connected shape. In our model (2) the range of \({\varvec{\Uptheta}}\) has been set in such a way that connectivity is lost for only a few extreme deformations. When this happens the connectivity may be recovered by means of standard morphological operations such as the Bridge and Skeletonisation [35]. However, these standard morphological operators have the tendency to smooth the original shape as the following example illustrates. Figure 9a shows a single deformation of T 0 corresponding to \({\varvec{\Uptheta}}=\left(0.4;1,-1\right).\) This represents one of the most extreme transformations that can be obtained by a single deformation. In order to recover a simply connected shape, first we apply a morphological Bridge transformation; see Fig. 9b. Although the shape is now connected, it is not simply connected since, the Bridge filter connects the borders every time the distance between them is less than two pixels so making narrow shapes multiply connected. One way to restore simple connectivity is to fill the internal holes and then to apply the morphological Skeletonisation; see Fig. 9c. This shape is now simply connected but it is much smoother than the original one in Fig. 9a with the result that some detailed information about the border has been lost. In order to obtain a more satisfactory connected shape than above we developed a new filter. Our new filter can be described as follows:
-
1.
Following a fixed sweeping scheme, all the image pixels are visited until a shape pixel is found. This pixel becomes the current pixel and forms the first pixel of the filtered shape.
-
2.
The second order neighbours of the current pixel are considered following a particular visiting scheme. The first shape pixel that has not yet been visited is selected. The current pixel becomes the old pixel and the selected pixel becomes the current pixel. The current pixel is added to the pixel of the filtered shape. The visiting scheme depends on the curvature of the shape at the current pixel and this is estimated by the relative location of the current and old pixel. Some visiting schemes are reported in Fig. 10. For the starting pixel of Step 1 a fixed visiting scheme is assumed.
-
3.
Step 2 is repeated until the current pixel is a neighbour of the starting pixel. This always happens since the shape obtained by applying the Bridge filter is connected and the direction (in our case anticlockwise) is maintained throughout the algorithm.
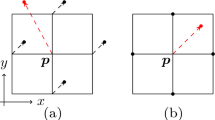
Visiting scheme for neighbouring pixels. The visiting order depends on the local curvature, estimated by the relative position of the current pixel ( cur ) and the old pixel ( old ). Three examples are shown. The first visiting scheme is the one adopted for the starting pixel, except in that case old would be replaced by 8
We call our new filter as the Ext filter. The result of this filter will always be the same when it is applied after the Bridge filter, provided that the starting shape pixel is one of the pixels of the outer border. In Fig. 9d we show the shape recovered by this filter. We note that our Ext filter has preserved the features of the border of the original shape shown in Fig. 9a much better than the previous approach shown in Fig. 9c. The Ext filter is also as fast as the morphology based approach (typically 1.6 × 10−2 s per reconstruction), and is implemented in one step instead of two.
Rights and permissions
About this article
Cite this article
de Pasquale, F., Stander, J. A multi-scale template method for shape detection with bio-medical applications. Pattern Anal Applic 12, 179–192 (2009). https://doi.org/10.1007/s10044-008-0114-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10044-008-0114-1