Abstract
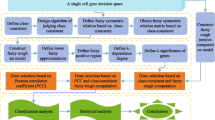
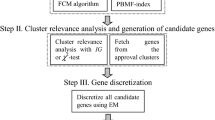
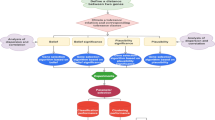
With the development of data mining, artificial intelligence, neural network, expert system and machine learning, information system (i-system) becomes more and more important. If the objects, attributes and information values in an i-system are replaced by cells, genes and gene expression values, respectively, then the i-system is said to be a gene space. Because gene expression data is characterized by small samples, high dimension and noise, there is considerable uncertainty in a gene space. Traditional machine learning and statistical methods are often powerless to a gene space. Granular computing (GrC) can effectively deal with various uncertainties. This paper studies the uncertainty measurement of gene space based on the class-consistent technology and discusses its application in gene selection from the perspective of GrC. A class-consistent relation between cells in a gene space is first established by the gene expression values of cells on the basis of class-consistent technology. Then, the information granules (i-granules) are obtained from a gene space by using the class-consistent relation. Next, two metrics (information granularity and information entropy) to measure the uncertainty of gene space are defined and their properties are also investigated. The results of numerical experiments and statistical tests verify their effectiveness. Furthermore, as their application to gene space, two gene selection algorithms are proposed. Finally, the clustering experiments and statistical tests on 16 gene spaces show that the designed gene selection algorithms outperform some state-of-the-art feature selection algorithms in terms of three clustering performance indicators.









Similar content being viewed by others
References
Alexander I, Tapani R (2010) Practical approaches to principal component analysis in the presence of missing values. J Mach Learn Res 11:1957–2000
Biase FH, Cao X, Zhong S (2014) Cell fate inclination within 2-cell and 4-cell mouse embryos revealed by single-cell RNA sequencing. Genome Res 24:1787–1796
Bommert A, Welchowski T, Schmid M, Rahnenführer J (2022) Benchmark of filter methods for feature selection in high-dimensional gene expression survival data. Briefings in Bioinformatics 23:bbab354
Cament L A, Castillo L E, Perez JP, Galdames FJ, Perez CA (2014) Fusion of local normalization and Gabor entropy weighted features for face identification. Pattern Recognit 47(2):568–577
Chung W, Eum HH, Lee HO, Lee KM, Lee HB, Kim KT, Ryu HS, Kim S, Lee JE, Park YH, Kan Z, Han W, Park WY (2017) Single-cell RNA-seq enables comprehensive tumour and immune cell profling in primary breast cancer. Nat Commun 8:15081
Dai JH, Hu H, Wu WZ, Qian YH, Huang DB (2018) Maximal discernibility-pair-based approach to attribute reduction in fuzzy rough sets. IEEE Trans Fuzzy Syst 26(4):2175–2187
Dai JH, Hu H, Zheng GJ, Hu QH, Han HF, Shi H (2016) Attribute reduction in interval-valued information systems based on information entropies. Front Inf Technol Electron Eng 17(9):919–928
Delgado A, Romero I (2016) Environmental conflict analysis using an integrated grey clustering and entropy-weight method: a case study of a mining project in Peru. Environ Model Softw 77:108–121
Dai JH, Tian HW (2013) Entropy measures and granularity measures for set valued information systems. Inf Sci 240:72–82
Dai JH, Wang WT, Xu Q (2013) An uncertainty measure for incomplete decision tables and its applications. IEEE Trans Cybern 43(4):1277–1289
Deng Q, Ramskld D, Reinius B, Sandberg R (2014) Single-cell RNA-seq reveals dynamic, random monoallelic gene expression in mammalian cells. Science 343:193–196
Engel I, Seumois G, Chavez L, Samaniego-Castruita D, White B, Chawla A, Mock D, Vijayanand P, Kronenberg M (2016) Innatelike functions of natural killer T cell subsets result from highly divergent gene programs. Nat Immunol 17:728–739
Fujita H, Gaeta A, Loia V, Orciuoli F (2019) Resilience analysis of critical infrastructures: a cognitive approach based on granular computing. IEEE Trans Cybern 49(5):1835–1848
Huang ZH, Li JJ Discernibility measures for fuzzy β-covering and their application, IEEE Transactions on Cybernetics
Goolam M, Scialdone A, Graham SJ, Macaulay IC, Jedrusik A, Hupalowska A, Voet T, Marioni JC, Zernicka-Goetz M (2016) Heterogeneity in Oct4 and Sox2 targets biases cell fate in 4-cell mouse embryos. Cell 165:61–74
Hu M, Tsang ECC, Guo YT, Xu WH Fast and robust attribute reduction based on the separability in fuzzy decision systems. IEEE Transactions on Cybernetics
Hempelmann CF, Sakoglu U, Gurupur VP, Jampana S (2016) An entropy-based evaluation method for knowledge bases of medical information systems. Expert Syst Appl 46:262–273
Hu QH, Yu DR, Liu J, Wu C (2008) Neighborhood rough set based heterogeneous feature subset selection. Information Sciences 178(18):3577–3594
Kolodziejczyk AA, Kim JK, Tsang JC, Ilicic T, Henriksson J, Natarajan KN, Tuck AC, Gao X, Buhler M, Liu P, Marioni JC, Teichmann SA (2015) Single cell RNA-sequencing of pluripotent states unlocks modular transcriptional variation. Cell Stem Cell 17:471–485
Li ZW, Liu XF, Dai JH, Chen JL, Fujita H (2020) Measures of uncertainty based on Gaussian kernel for a fully fuzzy information system. Knowl-Based Syst 196:105791
Liu KY, Li TY, Yang XB, Yang X, Liu D, Zhang PF, Wang J (2022) Granular cabin: An efficient solution to neighborhood learning in big data. Inf Sci 583:189–201
Liu KY, Yang XB, Fujita H, Liu D, Qian YH (2019) An efficient selector for multi-granularity attribute reduction. Inf Sci 505:457–472
Liu KY, Yang XB, Yu HL, Mi JS, Wang PX (2019) Rough set based semi-supervised feature selection via ensemble selector. Knowl-Based Syst 165:282–296
Li ZW, Zhang PF, Ge X, Xie NX, Zhang GQ, Wen CF (2019) Uncertainty measurement for a fuzzy relation information system. IEEE Trans Fuzzy Syst 27:2338–2352
Patel AP, Tirosh I, Trombetta JJ, Shalek AK, Gillespie SM, Wakimoto H, Cahill DP, Nahed BV, Curry WT, Martuza RL, Louis DN, Rozenblatt O, Suva ML, Regev A, Bernstein BE (2014) Single-cell rna-seq highlights intratumoral heterogeneity in primary glioblastoma. Science 344:401–1396
Pawlak Z (1991) Rough sets: Theoretical aspects of reasoning about data. Kluwer Academic Publishers, Dordrecht
Pollen AA, Nowakowski TJ, Shuga J, Wang XH, Leyrat AA, Lui JH, Li N, Szpankowski L, Fowler B, Chen P, Ramalingam N, Sun G, Thu M, Norris M, Lebofsky R, Toppani D, Kemp DW, Wong M, Clerkson B, Jones BN, Wu S, Knutsson L, Alvarado B, Wang J, Weaver LS, May AP, Jones RC, Unger MA, Kriegstein AR, West JA (2014) Low-coverage single-cell mRNA sequencing reveals cellular heterogeneity and activated signaling pathways in developing cerebral cortex. Nat Biotechnol 32:1053–1058
Qian YH, Liang JY, Wu WZ, Dang CY (2011) Information granularity in fuzzy binary GrC model. IEEE Trans Fuzzy Syst 19:253–264
Ramskold D, Luo S, Wang YC, Li R, Deng Q, Faridani OR, Daniels GA, Khrebtukova I, Loring JF, Laurent LC, Schroth GP, Sandberg R (2012) Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells. Nat Biotech 30:777–782
Robert JK, Lee G, Li JW, Genshaft AS, Kazer SW, Payer KR, Borrajo J, Blainey PC, Irvine DJ, Shalek AK, Manalis SR (2016) A microfuidic platform enabling single-cell RNA-seq of multigenerational lineages. Nat Commun 7:10220
Song Y, Botvinnik OB, Lovci MT, Kakaradov B, Liu P, Xu JL, Yeo GW (2017) Single-cell alternative splicing analysis with expedition reveals splicing dynamics during neuron diferentiation. Molecular Cell 67:148–161
Song QJ, Jiang HY, Liu J (2017) Feature selection based on FDA and F-score for multi-class classification. Expert Syst Appl 81:22–27
Sheng J, Li WV (2021) Selecting gene features for unsupervised analysis of single-cell gene expression data. Briefings in Bioinformatics 22:bbab295
Sasagawa Y, Nikaido T, Hayashi T, Danno H, Uno KD, Imai T, Ueda HR (2013) Quartz-Seq: a highly reproducible and sensitive single-cell RNA sequencing method, reveals nongenetic geneexpression heterogeneity. Genome Biology 14:3097
Sharma A, Lysenko A, Boroevich K A, Vans E, Tsunoda T (2021) DeepFeature: feature selection in non-image data using convolutional neural network. Briefings in Bioinformatics 22:bbab297
Sharma A, Rani R (2019) C-HMOSHSSA: Gene selection for cancer classification using multi-objective meta-heuristic and machine learning methods. Comput Methods Programs Biomed 178:219–235
Saqlain SM, Sher M, Shah FA, Khan I, Ashraf MU, Awais M, Ghani A (2019) Fisher score and Matthews correlation coefficient-based feature subset selection for heart disease diagnosis using support vectormachines. Knowl Inf Syst 58:139–167
Singh S, Shreevastava S, Som T, Somani G (2020) A fuzzy similarity-based rough set approach for attribute selection in set-valued information systems. Soft Comput 24:4675–4691
Sun L, Zhang XY, Qian YH, Xu JC, Zhang SG, Tian Y (2019) Joint neighborhood entropy-based gene selection method with fisher score for tumor classification. Appl Intell 49:1245–1259
Ting DT, Wittner BS, Ligorio M, Jordan NV, Shah AM, Miyamoto DT, Aceto N, Bersani F, Brannigan BW, Xega K, Ciciliano JC, Zhu H, MacKenzie OC, Trautwein J, Arora KS, Shahid M, Ellis HL, Qu N, Haber DA, Single-cell R N A (2014) sequencing identifies extracellular matrix gene expression by pancreatic circulating tumor cells. Cell Reports 8:1905– 1918
Treutlein B, Brownfeld DG, Wu AR, Nef NF, Mantalas GL, Espinoza FH, Desai TJ, Krasnow MA, Quake SR (2014) Reconstructing lineage hierarchies of the distal lung epithelium using single-cell RNA-seq. Nature 509:371–375
Tan AH, Shi SW, Wu WZ, Li JJ, Pedrycz W. Granularity and entropy of intuitionistic fuzzy information and their applications. IEEE Transactions on Cybernetics
Usoskin D, Furlan A, Islam S, Abdo H, Lnnerberg P, Lou D, Hjerling J, Haeggstrm J, Kharchenko O, Kharchenko PV, Linnarsson S, Ernfors P (2015) Unbiased classifcation of sensory neuron types by large-scale single-cell RNA sequencing. Nat Neurosci 18:53–145
Wang YB, Chen XJ, Dong K (2019) Attribute reduction via local conditional entropy. Int J Mach Cybern 10(12):3619–3634
Wang CZ, Huang Y, Shao MW, Hu QH, Chen DG (2020) Feature selection based on neighborhood self-information. IEEE Trans Cybern 50(9):4031–4042
Wu Y, Zhang K (2020) Tools for the analysis of high-dimensional single-cell RNA sequencing data. Nat Rev Nephrol 16:408–421
Xie SD, Wang YX (2014) Construction of tree network with limited delivery latency in homogeneous wireless sensor networks. Wirel Pers Commun 78(1):231–246
Yan L, Yang M, Guo H, Yang L, Wu J, Li R, Liu P, Lian Y, Zheng X, Yan J, Huang J, Li M, Wu X, Wen L, Lao K, Li R, Qiao J, Tang F (2013) Single-cell RNA-Seq profling of human preimplantation embryos and embryonic stem cells. Nat Struct Mol Biol 20:1131–1139
Yang Y, Huh R, Houston WC, Lin Y, Michael IL, Li Y (2019) SAFE-clustering: Single-cell Aggregated (from Ensemble) clustering for single-cell RNA-seq data. Bioinformatics 35:1269–1277
Zadeh LA (1997) Toward a theory of fuzzy information granulation and its centrality in human reasoning and fuzzy logic. Fuzzy Sets Syst 90:111–127
Zhang GQ, Li ZW, Wu WZ, Liu XF, Xie NX (2018) Information structures and uncertainty measures in a fully fuzzy information system. Int J Approx Reason 101:119–149
Acknowledgements
The authors would like to thank the editors and the anonymous reviewers for their valuable comments and suggestions, which have helped immensely in improving the quality of the paper. This work is supported by Doctoral Research Start Project (CZ2021YJRC01), Natural Science Foundation of Guangxi (2020GXNSFAA159155, AD19245102) and Special Scientific Research Project of Young Innovative Talents in Guangxi (2019AC20052).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Li, Z., Zhang, Q., Wang, P. et al. Uncertainty measurement for a gene space based on class-consistent technology: an application in gene selection. Appl Intell 53, 5416–5436 (2023). https://doi.org/10.1007/s10489-022-03657-3
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10489-022-03657-3