Abstract
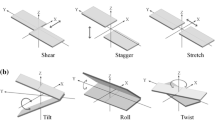
Non-canonical base pairs contribute immensely to the structural and functional variability of RNA, which calls for a detailed characterization of their spatial conformation. Intra-base pair parameters, namely propeller, buckle, open-angle, stagger, shear and stretch describe structure of base pairs indicating planarity and proximity of association between the two bases. In order to study the conformational specificities of non-canonical base pairs occurring in RNA crystal structures, we have upgraded NUPARM software to calculate these intra-base pair parameters using a new base pairing edge specific axis system. Analysis of base pairs and base triples with the new edge specific axis system indicate the presence of specific structural signatures for different classes of non-canonical pairs and triples. Differentiating features could be identified for pairs in cis or trans orientation, as well as those involving sugar edges or C–H-mediated hydrogen bonds. It was seen that propeller for all types of base pairs in cis orientation are generally negative, while those for trans base pairs do not have any preference. Formation of a base triple is seen to reduce propeller of the associated base pair along with reduction of overall flexibility of the pairs. We noticed that base pairs involving sugar edge are generally more non-planar, with large propeller or buckle values, presumably to avoid steric clash between the bulky sugar moieties. These specific conformational signatures often provide an insight into their role in the structural and functional context of RNA.





Similar content being viewed by others
References
Gesteland RF, Cech TR, Atkins JF (1999) The RNA world, 2nd edn. Cold Spring Harbor Laboratory, New York
Hermann T, Westhof E (1999) Chem Biol 6:R335
Hoogsteen K (1963) Acta Crystallogr 16:907
Topal MD, Fresco JR (1976) Nature 263:289
Wahl MC, Rao ST, Sundaralingam M (1996) Nat Struct Biol 3:24
Nissen P, Ippolito JA, Ban N, Moore PB, Steitz TA (2001) Proc Natl Acad Sci USA 98:4899
Leontis NB, Westhof E (2001) RNA 7:499
Leontis NB, Stombaugh J, Westhof E (2002) Nucleic Acids Res 30:3497
Duarte CM, Pyle AM (1998) J Mol Biol 284:1465
Gendron P, Lemieux S, Major F (2001) J Mol Biol 308:919
Klosterman PS, Tamura M, Holbrook SR, Brenner SE (2002) Nucleic Acids Res 30:392
Lemieux S, Major F (2002) Nucleic Acids Res 30:4250
Nagaswamy U, Larios-Sanz M, Hury J, Collins S, Zhang Z, Zhao Q, Fox GE (2002) Nucleic Acids Res 30:395
Walberer BJ, Cheng AC, Frankel AD (2003) J Mol Biol 327:767
Lee JC, Gutell RR (2004) J Mol Biol 344:1225
Sykes MT, Levitt M (2005) J Mol Biol 351:26
Das J, Mukherjee S, Mitra A, Bhattacharyya D (2006) J Biomol Struct Dyn 24:149
Saenger W (1984) Principles of nucleic acid structure. Springer, Berlin Heidelberg New York
Sponer JE, Spackova N, Kulhanek P, Leszczynski J, Sponer J (2005) J Phys Chem 109A:2292
Bhattacharyya D, Koripella SC, Mitra A, Rajendran VB, Sinha B (2006) Manuscript in preparation
Mathews DH, Sabina J, Zuker M, Turner DH (1999) J Mol Biol 288:911
Dima RI, Hyeon C, Thirumalai D (2005) J Mol Biol 347:53
Florian J, Sponer J, Warshel A (1999) J Phys Chem B 103:884
Go M, Go N (1976) Biopolymers 15:1119
Olson WK, Gorin AA, Lu XJ, Hock LM, Zhurkin VB (1998) Proc Natl Acad Sci USA 95:11163
Olson WK, Bansal M, Burley SK, Dickerson RE, Gerstein M, Harvey SC, Heinmann U, Lu XJ, Neidle S, Shakked Z, Sklenar H, Suzuki M, Tung CS, Westhof E, Wolberger C, Berman HM (2001) J Mol Biol 313:229
Calladine CR (1982) J Mol Biol 161:343
Bhattacharyya D, Bansal M (1992) J Biomol Struct Dyn 10:213
Lu XJ, Olson WK (2003) Nucleic Acids Res 31:5108
Leontis NB, Westhof E (2003) Curr Opin Struct Biol 13:300
Stark A, Brennecke J, Russell RB, Cohen SM (2003) PloS Biol 1:397
Peyret N, Seneviratne A, Allawi HT, SantaLucia J Jr (1999) Biochemistry 38:3468
Michel F, Westhof E (1990) J Mol Biol 216:585
Gautheret D, Damberger SH, Gutell RR (1995) J Mol Biol 248:27
Gautheret D, Gutell RR (1997) Nucleic Acids Res 25:1559
Razga F, Koca J, Sponer J, Leontis NB (2005) Biophys J 88:3466
Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE (2000) Nucleic Acids Res 28:235
Yang H, Jossinet F, Leontis N, Chen L, Westbrook J, Berman H, Westhof E (2003) Nucleic Acids Res 31:3450
Berman HM, Olson WK, Beveridge DL, Westbrook J, Gelbin A, Demeny T, Hsieh SH, Srinivasan AR, Schneider B (1992) Biophys J 63:751
El Hassan MA, Calladine CR (1995) J Mol Biol 251:648
Lu XJ, El Hassan MA, Hunter CA (1997) J Mol Biol 273:668
Gorin AA, Zhurkin VB, Olson WK (1995) J Mol Biol 247:34
Kosikov KM, Gorin AA, Zhurkin VB, Olson WK (1999) J Mol Biol 289:1301
Lavery R, Sklenar H (1988) J Biomol Struct Dyn 6:63
Lavery R, Sklenar H (1989) J Biomol Struct Dyn 6:655
Dickerson RE (1998) Nucleic Acids Res 26:1906
Soumpasis DM, Tung CS (1988) J Biomol Struct Dyn 6:397
Tung CS, Soumpasis DM, Hummer G (1994) J Biomol Struct Dyn 11:1327
Bansal M, Bhattacharyya D, Ravi B (1995) CABIOS 11:281
Bhattacharyya D, Bansal M (1989) J Biomol Struct Dyn 6:645
Babcock MS, Pednault EPD, Olson WK (1993) J Biomol Struct Dyn 11:597
Babcock MS, Pednault EPD, Olson WK (1994) J Mol Biol 237:125
Babcock MS, Olson WK (1994) J Mol Biol 237:98
Holbrook SR (2005) Curr Opin Struct Biol 15:302
Ghosh A, Bansal M (1999) J Mol Biol 294:1149
Bandyopadhyay D, Bhattacharyya D (2006) Bioploymers 83:313
Acknowledgments
We are grateful to the Council of Scientific and Industrial Research (CSIR) and Department of Biotechnology (DBT), India, for financial support. We are thankful to Ms Jhuma Das for technical support.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Mukherjee, S., Bansal, M. & Bhattacharyya, D. Conformational specificity of non-canonical base pairs and higher order structures in nucleic acids: crystal structure database analysis. J Comput Aided Mol Des 20, 629–645 (2006). https://doi.org/10.1007/s10822-006-9083-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10822-006-9083-x