Abstract
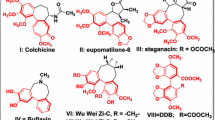
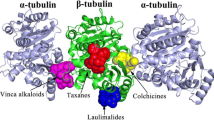
Noscapine and its derivatives bind stoichiometrically to tubulin, alter its dynamic instability and thus effectively inhibit the cellular proliferation of a wide variety of cancer cells including many drug-resistant variants. The tubulin molecule is composed of α- and β-tubulin, which exist as various isotypes whose distribution and drug-binding properties are significantly different. Although the noscapinoids bind to a site overlapping with colchicine, their interaction is more biased towards β-tubulin. In fact, their precise interaction and binding affinity with specific isotypes of β-tubulin in the αβ-heterodimer has never been addressed. In this study, the binding affinity of a panel of noscapinoids with each type of tubulin was investigated computationally. We found that the binding score of a specific noscapinoid with each type of tubulin isotype is different. Specifically, amino-noscapine has the highest binding score of −6.4, −7.2, −7.4 and −7.3 kcal/mol with αβI, αβII, αβIII and αβIV isotypes, respectively. Similarly 10 showed higher binding affinity of −6.8 kcal/mol with αβV, whereas 8 had the highest binding affinity of −7.2, −7.1 and −7.2 kcal/mol, respectively with αβVI, αβVII and αβVIII isotypes. More importantly, both amino-noscapine and its clinical derivative, bromo-noscapine have the highest binding affinity of −46.2 and −38.1 kcal/mol against αβIII (overexpression of αβIII has been associated with resistance to a wide range of chemotherapeutic drugs for several human malignancies) as measured using MM-PBSA. Knowledge of the isotype specificity of the noscapinoids may allow for development of novel therapeutic agents based on this class of drugs.






Similar content being viewed by others
References
Stanton RA, Gernert KM, Nettles JH, Aneja R (2011) Drugs that target dynamic microtubules: a new molecular perspective. Med Res Rev 31(3):443–481
Checchi PM, Nettles JH, Zhou J, Snyder JP, Joshi HC (2003) Microtubule-interacting drugs for cancer treatment. Trends Pharmacol Sci 24(7):361–365
Downing KH (2000) Structural basis for the interaction of tubulin with proteins and drugs that affect microtubule dynamics. Annu Rev Cell Dev Biol 16:89–111
Alam MA, Naik PK (2009) Molecular modelling evaluation of the cytotoxic activity of podophyllotoxin analogues. J Comput Aided Mol Des 23:209–225
Luduena RF (1998) Multiple forms of tubulin: different gene products and covalent modifications. Int Rev Cytol 178:207–275
Banerjee A, Luduena RF (1992) Kinetics of colchicine binding to purified beta tubulin isotypes from bovine brain. J Biol Chem 267:13335–13339
Mozzetti S, Ferlini C, Concolino P et al (2005) Class III B-tubulin overexpression is a prominent mechanism of paclitaxel resistance in ovarian cancer patients. Clin Cancer Res 11:298–305
Joshua A, McCarroll Gan PP, Liu M, Kavallaris M (2010) βIII-tubulin is a multifunctional protein involved in drug sensitivity and tumorigenesis in non-small cell lung cancer. Clin Cancer Res 70(12):4995–5003
Nicoletti MI, Valoti G, Giannakakou P et al (2001) Expression of β_tubulin isotypes in human ovarian carcinoma xenografts and in a sub-panel of human cancer cell lines from the NCI-anticancer drug screen: correlation with sensitivity to microtubule active agents. Clin Cancer Res 7:2912–2922
Zhang Y, Yang H, Liu J et al (2013) High expression levels of class III β-tubulin in resected non-small cell lung cancer patients are predictive of improved patient survival after vinorelbine-based adjuvant chemotherapy. Oncol Lett 6:220–226
Huzil JT, Luduena RF, Tuszynski J (2006) Comparative modelling of human beta-tubulin isotypes and implications for drug binding. Nanotechnology 17:S90–S100
Mane JY, Klobukowski MJ, Huzil T, Tuszynski J (2008) Free energy calculations on the binding of colchicine and its derivatives with the α/β-tubulin isoforms. J Chem Inf Model 48(9):1824–1832
Gan PP, Pasquier E, Kavallaris M (2000) Class III beta-tubulin mediates sensitivity to chemotherapeutic drugs in non small cell lung cancer. Cancer Res 67:9356–9363
Kim YS, Tseng CY, Mane JY, Winter P, Johnson L, Huzil T, Izbicka E, Luduena RF, Tuszynski J (2010) Quantitative analysis of the effect of tubulin isotype expression on sensitivity of cancer cell lines to a set of novel colchicine derivatives. Mol Cancer 9:131
Christoph DC, Kasper S, Gauler TC, Loesch C, Engelhard M, Theegarten D, Poettgen C, Hepp R, Peglow A, Loewendick H, Welter S, Stamatis G, Hirsch FR, Schuler M, Eberhardt WE, Wohlschlaeger J (2012) βV-tubulin expression is associated with outcome following taxane-based chemotherapy in non-small cell lung cancer. Br J Cancer 107:823–830
Ye K, Ke Y, Keshava N, Shanks J, Kapp JA, Tekmal RR, Petros J, Joshi HC (1998) Opium alkaloid noscapine is an antitumor agent that arrests metaphase and induces apoptosis in dividing cells. Proc Natl Acad Sci USA 95:1601–1606
Joshi HC, Salil A, Bughani U, Naik PK (2010) Noscapinoids: a new class of anticancer drugs demand bio-technological intervention. In: Arora R (ed) Medicinal plant biotechnology, CAB e-Books CABI (H ISBN 9781845936785)
Mahmoudian M, Rahimi-Moghaddam P (2009) The anti-cancer activity of noscapine: a review. Recent Pat Anticancer Drug Discov 4(1):92–97
Landen J, Lang WR, McMahon SJ, Rusan NM, Yvon AM, Adams AW, Sorcinelli MD, Campbell R, Bonaccorsi P, Ansel JC, Archer DR, Wadsworth P, Armstrong CA, Joshi HC (2002) Noscapine alters microtubule dynamics in living cells and inhibits the progression of melanoma. Cancer Res 62:4109–4114
Zhou J, Panda D, Landen JW, Wilson L, Joshi HC (2002) Minor alteration of microtubule dynamics causes loss of tension across kinetochore pairs and activates the spindle checkpoint. J Biol Chem 277:17200–17208
Aneja R, Vangapandu SN, Lopus M, Viswesarappa VG, Dhiman N, Verma A, Chandra R, Panda D, Joshi HC (2006) Synthesis of microtubule-interfering halogenated noscapine analogues that perturb mitosis in cancer cells followed by cell death. Biochem Pharmacol 72:415–426
Zhou J, Gupta K, Aggarwal S, Aneja R, Chandra R, Panda D, Joshi HC (2003) Brominated derivatives of noscapine are potent microtubule-interfering agents that perturb mitosis and inhibit cell proliferation. Mol Pharmacol 63:799–807
Aneja R, Vangapandu SN, Lopus M, Chandra R, Panda D, Joshi HC (2006) Development of a novel nitro-derivative of Noscapine for the potential treatment of drug-resistant ovarian cancer and Tcell lymphoma. Mol Pharmacol 69:1801–1809
Naik PK, Chatterji BP, Vangapandu SN, Aneja R, Chandra R, Kanteveri S, Joshi HC (2011) Rational design synthesis and biological evaluations of amino-noscapine: a high affinity tubulin-binding noscapinoids. J Comput Aided Drug Des 25:443–454
Zhou J, Gupta K, Yao J, Ye K, Panda D, Giannakakou P, Joshi HC (2002) Paclitaxel-resistant human ovarian cancer cells undergo v-Jun NH2-terminl kinase-mediated apoptosis in response to noscapine. J Biol Chem 277(42):39777–39785
Santoshi S, Naik PK, Joshi HC (2011) Rational design of novel anti-microtubule agent (9-azido-noscapine) from quantitative structure activity relationship (QSAR) evaluation of noscapinoids. J Biomol Screen 16:1047–1058
Manchukonda NK, Naik PK, Santoshi S, Lopus M, Joseph S, Sridhar B, Kantevari S (2013) Rational design synthesis and biological evaluation of third generation α-noscapine analogues as potent tubulin binding anti-cancer agents. PLoS ONE 8(10):77970
Naik PK, Santoshi S, Rai A, Joshi HC (2011) Molecular modelling and competition binding study of Br-noscapine and colchicine provide insight into noscapinoid-tubulin binding site. J Mol Graph Model 29:947–955
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Berendsen HJC, Spoel VD, Drunen VR (1995) GROMACS: a message passing parallel molecular dynamics implementation. Comput Phys Commun 91:43–56
Daren T, York D, Pedersen L (1993) Particle mesh Ewald: an N-log(N) method for Ewald sums in large systems. J Chem Phys 98:10089–10092
Hess B, Bekker H, Berendsen HJC, Fraaije JGEM (1997) LINCS: a linear constraint solver for molecular simulations. J Comput Chem 18:1463–1472
Laskowski RA, MacArthur MW, Moss DS, Thornton JM (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J App Crystal 26:283–291
Ramachandran GN, Ramakrishnan C, Sasisekharan V (1963) Stereochemistry of polypeptide chain configurations. J Mol Biol 7:95–99
Colovos C, Yeates TO (1993) Verification of protein structures: patterns of non-bonded atomic interactions. Protein Sci 2:1511–1519
Eisenberg D, Luthy R, Bowie JU (1997) VERIFY3D: assessment 694 of protein models with three-dimensional profiles. Methods Enzymol 277:396–404
Lee C, Yang W, Parr RG (1988) Development of the Colle-Salvetti correlation-energy formula into a functional of the electron density. Phys Rev B 37:785–789
Becke AD (1993) A new mixing of Hartree-Fock and local density-functional theories. J Chem Phys 98:1372–1377
Binkley JS, Pople JA, Hehre WJ (1980) Self-consistent molecular orbital methods 21 small split-valence basis sets for first-row elements. J Am Chem Soc 102:939–947
Gordon MS, Binkley JS, Pople JA, Pietro WJ, Hehre WJ (1982) Self-consistent molecular-orbital methods 22 small split-valence basis sets for second-row elements. J Am Chem Soc 104:2797–2803
Pietro WJ, Francl MM, Hehre WJ, Defrees DJ, Pople JA, Binkley JS (1982) Self-consistent molecular orbital methods 24 supplemented small split-valence basis sets for second-row elements. J Am Chem Soc 104:5039–5048
Friesner RA, Banks JL, Murphy RB, Halgren TA, Klicic JJ, Mainz DT, Repasky MP, Knoll EH, Shelley M, Perry JK, Shaw DE, Francis P, Shenkin PS (2004) Glide: a new approach for rapid accurate docking and scoring 1 method and assessment of docking accuracy. J Med Chem 47:1739–1749
Halgren TA, Murphy RB, Friesner RA, Beard HS, Frye LL, Pollard WT, Banks JL (2004) Glide: a new approach for rapid accurate docking and scoring 2 enrichment factors in database screening. J Med Chem 47:1750–1759
Case DA, Walker RC et al (2010) AMBER 11. University of California, San Francisco
Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz KM Jr, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA (1995) A second generation force field for the simulation of proteins nucleic acids and organic molecules. J Am Chem Soc 117:5179–5197
Wang JM, Wolf RM, Caldwell JW, Kollamn PA, Case DA (2004) Development and testing of a general amber force field. J Comput Chem 25:1157–1174
Ryckaert JP, Ciccotti G, Berendsen HJC (1977) Numerical integration of the Cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J Comput Phys 23:327–341
Hornak V, Abel R, Okur A, Strockbine B, Roitberg A, Simmerling C (2006) Comparison of multiple Amber force fields and development of improved protein backbone parameters. Protein 65:712–725
Berendsen HJC, Postma JP, van Gunsteren WF, DiNola A, Haak JR (1984) Molecular dynamics with coupling to an external bath. J Chem Phys 81:3684–3691
Kollman PA, Massova I, Reyes C, Kuhn B, Huo S, Chong L, Lee M, Lee T, Duan Y, Wang W, Donini O, Cieplak P, Srinivasan J, Case DA, Cheatham TE (2000) Calculating structures and free energies of complex molecules: combining molecular mechanics and continuum models. Acc Chem Res 33:889–897
Shen XL, Kamimura MT, Wei J, Gao QZ (2012) Computer-aided de novo ligand design and docking/molecular dynamics study of vitamin D receptor agonists. J Mol Model 18:203–212
Niu X, Gao X, Wang H, Wang X, Wang S (2013) Insight into the dynamic interaction between different flavonoids and bovine serum albumin using molecular dynamics simulations and free energy calculations. J Mol Model 19:1039–1047
Guo J, Wang X, Sun H, Liu H, Yao X (2012) T he molecular basis of IGF-II/IGF2R recognition: acombined molecular dynamics simulation free-energy calculation and computational alanine scanning study. J Mol Model 18:1421–1430
Acknowledgments
Authors are thankful to Dr. Harish Joshi, School of Medicine, Emory University, Georgia, USA, for carefully reading the manuscript and for providing constructive criticism. We are greatly indebted to the anonymous reviewers of this manuscript for their helpful suggestions.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Santoshi, S., Naik, P.K. Molecular insight of isotypes specific β-tubulin interaction of tubulin heterodimer with noscapinoids. J Comput Aided Mol Des 28, 751–763 (2014). https://doi.org/10.1007/s10822-014-9756-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10822-014-9756-9