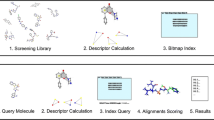
Abstract
In many practical applications of structure-based virtual screening (VS) ligands are already known. This circumstance requires that the obtained hits need to satisfy initial made expectations i.e., they have to fulfill a predefined binding pattern and/or lie within a predefined physico-chemical property range. Based on the RApid Index-based Screening Engine (RAISE) approach, we introduce cRAISE—a user-controllable structure-based VS method. It efficiently realizes pharmacophore-guided protein-ligand docking to assess the library content but thereby concentrates only on molecules that have a chance to fulfill the given binding pattern. In order to focus only on hits satisfying given molecular properties, library profiles can be utilized to simultaneously filter compounds. cRAISE was evaluated on a range of strict to rather relaxed hypotheses with respect to its capability to guide binding-mode predictions and VS runs. The results reveal insights into a guided VS process. If a pharmacophore model is chosen appropriately, a binding mode below 2 Å is successfully reproduced for 85 % of well-prepared structures, enrichment is increased up to median AUC of 73 %, and the selectivity of the screening process is significantly enhanced leading up to seven times accelerated runtimes. In general, cRAISE supports a versatile structure-based VS approach allowing to assess hypotheses about putative ligands on a large scale.










Similar content being viewed by others
References
Sotriffer C (2011) Virtual screening. WILEY-VCH Verlag GmbH & Co, KGaA, Weinheim
Sotriffer C, Matter H (2011) The challenge of affinity prediction: scoring functions for structure-based virtual screening. In: Sotriffer C (ed) Virtual screening. Wiley-VCH Verlag GmbH & Co, KGaA, Weinheim, pp 177–221
Henzler AM, Rarey M (2010) Mol Inform 29:164–173
Sanders MP, McGuire R, Roumen L, de Esch IJ, de Vlieg J, Klomp JP, de Graaf C (2012) Med Chem Commun 3:28–38
Wallach I (2011) Drug Dev Res 72:17–25
Leach AR, Gillet VJ, Lewis RA, Taylor R (2010) J Med Chem 53:539–558
Yang SY (2010) Drug Discov Today 15:444–450
Dror O, Shulman-Peleg A, Nussinov R, Wolfson H (2004) Curr Med Chem 11:71–90
Tintori C, Corradi V, Magnani M, Manetti F, Botta M (2008) J Chem Inf Model 48:2166–2179
Muthas D, Sabnis YA, Lundborg M, Karln A (2008) J Mol Graph Model 26:1237–1251
Peach ML, Nicklaus MC (2009) J Chem Inform 1:6
Yang JM, Shen TW (2005) Proteins 59:205–220
Fradera X, Knegtel RM, Mestres J (2000) Proteins 40:623–636
Verdonk ML, Berdini V, Hartshorn MJ, Mooij WTM, Murray CW, Taylor RD, Watson P (2004) J Chem Inf Comp Sci 44:793–806
Hindle SA, Rarey M, Buning C, Lengauer T (2002) J Comput Aided Mol Des 16:129–149
Rarey M, Kramer B, Lengauer T, Klebe G (1996) J Mol Biol 261:470–489
Schlosser J, Rarey M (2009) J Chem Inf Model 49:800–809
Schellhammer I, Rarey M (2007) J Comput Aided Mol Des 21:223–238
Urbaczek S, Kolodzik A, Fischer JR, Lippert T, Heuser S, Groth I, Schulz-Gasch T, Rarey M (2011) J Chem Inf Model 51:3199–3207
Hartshorn MJ, Verdonk ML, Chessari G, Brewerton SC, Mooij WTM, Mortenson PN, Murray CW (2007) J Med Chem 50:726–741
Huang N, Shoichet BK, Irwin JJ (2006) J Med Chem 49:6789–6801
Neves MAC, Totrov M, Abagyan R (2012) J Comput Aided Mol Des 26:675–686
Spitzer R, Jain AN (2012) J Comput Aided Mol Des 26:687–699
Schneider N, Hindle S, Lange G, Klein R, Albrecht J, Briem H, Beyer K, Claußen H, Gastreich M, Lemmen C, Rarey M (2012) J Comput Aided Mol Des 26:701–723
Novikov FN, Stroylov VS, Zeifman AA, Stroganov OV, Kulkov V, Chilov GG (2012) J Comput Aided Mol Des 26:725–735
Liebeschuetz JW, Cole JC, Korb O (2012) J Comput Aided Mol Des 26:737–748
Brozell SR, Mukherjee S, Balius TE, Roe DR, Case DA, Rizzo RC (2012) J Comput Aided Mol Des 26:749–773
Corbeil CR, Williams CI, Labute P (2012) J Comput Aided Mol Des 26:775–786
Repasky MP, Murphy RB, Banks JL, Greenwood JR, Tubert-Brohman I, Bhat S, Friesner RA (2012) J Comput Aided Mol Des 26:787–799
Irwin JJ, Shoichet BK (2005) J Chem Inf Model 45:177–182
Irwin JJ, Sterling T, Mysinger MM, Bolstad ES, Coleman RG (2012) J Chem Inf Model 52:1757–1768
Schärfer C, Schulz-Gasch T, Hert J, Heinzerling L, Schulz B, Inhester T, Stahl M, Rarey M (2013) Chem Med Chem 8:1690–1700
Wu K (2005) J Phys: Conf Ser 16:556–560
Wu K, Ahern S, Bethel EW, Chen J, Childs H, Cormier-michel E, Geddes C, Gu J, Hagen H, Hamann B, Koegler W, Lauret J, Meredith J, Messmer P, Otoo E, Perevoztchikov V, Poskanzer A, Rübel O, Shoshani A, Sim E, Stockinger K, Weber G, Zhang Wming (2009) J Phys Conf Ser 180:1
Kirchmair J, Ristic S, Eder K, Markt P, Wolber G, Laggner C, Langer T (2007) J Chem Inf Model 47:2182–2196
Schulz-Gasch T, Schärfer C, Guba W, Rarey M (2012) J Chem Inf Model 52:1499–1512
Hilbig M, Urbaczek S, Groth I, Heuser S, Rarey M (2013) J Chem Inform 5:38
Barber CB, Dobkin DP, Huhdanpaa H (1996) ACM Trans Math Softw 22:469–483
Böhm HJ (1994) J Comput Aided Mol Des 8:243–256
Warren GL, Do TD, Kelley BP, Nicholls A, Warren SD (2012) Drug Discov Today 17:1270–1281
Zinc clean leads (2012) UCSF Univerity of California, San Francisco. http://zinc.docking.org/subsets/clean-leads. Accessed 7 Dec 2012
Acknowledgments
We would like to thank Nadine Schneider for the fruitful discussions about protein-ligand interactions and scoring functions, moreover, Christin Schärfer for her commitments to conformer generation. This work was financially supported by the BMWI-ZIM Project KF2563701.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Henzler, A.M., Urbaczek, S., Hilbig, M. et al. An integrated approach to knowledge-driven structure-based virtual screening. J Comput Aided Mol Des 28, 927–939 (2014). https://doi.org/10.1007/s10822-014-9769-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10822-014-9769-4