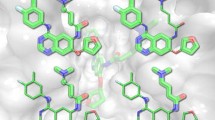
Abstract
Our group has recently demonstrated that virtual screening is a useful technique for the identification of target-specific molecular probes. In this paper, we discuss some of our proof-of-concept results involving two biologically relevant target proteins, and report the development of a computational script to generate large databases of fluorescence-labelled compounds for computer-assisted molecular design. The virtual screening of a small library of 1,153 fluorescently-labelled compounds against two targets, and the experimental testing of selected hits reveal that this approach is efficient at identifying molecular probes, and that the screening of a labelled library is preferred over the screening of base compounds followed by conjugation of confirmed hits. The automated script for library generation explores the known reactivity of commercially available dyes, such as NHS-esters, to create large virtual databases of fluorescence-tagged small molecules that can be easily synthesized in a laboratory. A database of 14,862 compounds, each tagged with the ATTO680 fluorophore was generated with the automated script reported here. This library is available for downloading and it is suitable for virtual ligand screening aiming at the identification of target-specific fluorescent molecular probes.









Similar content being viewed by others
Explore related subjects
Discover the latest articles and news from researchers in related subjects, suggested using machine learning.References
Johnson I, Michelle TZS (eds) (2010) Molecular probes handbook: a guide to fluorescent probes and labeling technologies, 11th edn. Life Technologies Coorporation, USA
Giepmans BN, Adams SR, Ellisman MH, Tsien RY (2006) The fluorescent toolbox for assessing protein location and function. Science (New York, NY) 312(5771):217–224. doi:10.1126/science.1124618
Riggs J, Seiwald R, Burckhalter J, Downs CM, Metcalf T (1958) Isothiocyanate compounds as fluorescent labeling agents for immune serum. Am J Pathol 34(6):1081
Lazarides E, Weber K (1974) Actin antibody: the specific visualization of actin filaments in non-muscle cells. Proc Natl Acad Sci 71(6):2268–2272
Sack U, Conrad K, Csernok E, Frank I, Hiepe F, Krieger T, Kromminga A, Von Landenberg P, Messer G, Witte T, Mierau R (2009) Autoantibody detection using indirect immunofluorescence on HEp-2 cells. Dtsch Med Wochenschr 134(24):1278–1282. doi:10.1055/s-0029-1225278
Hoxha E, Harendza S, Zahner G, Panzer U, Steinmetz O, Fechner K, Helmchen U, Stahl RAK (2011) An immunofluorescence test for phospholipase-A2-receptor antibodies and its clinical usefulness in patients with membranous glomerulonephritis. Nephrol Dial Transplant 26(8):2526–2532. doi:10.1093/ndt/gfr247
Kinders RJ, Hollingshead M, Lawrence S, Ji J, Tabb B, Bonner WM, Pommier Y, Rubinstein L, Evrard YA, Parchment RE, Tomaszewski J, Doroshow JH (2010) Development of a validated immunofluorescence assay for γH2AX as a pharmacodynamic marker of topoisomerase I inhibitor activity. Clin Cancer Res 16(22):5447–5457. doi:10.1158/1078-0432.ccr-09-3076
Schmidt E, Zillikens D (2010) Modern diagnosis of autoimmune blistering skin diseases. Autoimmun Rev 10(2):84–89. doi:10.1016/j.autrev.2010.08.007
Chames P, Van Regenmortel M, Weiss E, Baty D (2009) Therapeutic antibodies: successes, limitations and hopes for the future. Br J Pharmacol 157(2):220–233. doi:10.1111/j.1476-5381.2009.00190.x
Ntziachristos V, Bremer C, Weissleder R (2003) Fluorescence imaging with near-infrared light: new technological advances that enable in vivo molecular imaging. Eur Radiol 13(1):195–208. doi:10.1007/s00330-002-1524-x
Hilderbrand SA, Weissleder R (2010) Near-infrared fluorescence: application to in vivo molecular imaging. Curr Opin Chem Biol 14(1):71–79. doi:10.1016/j.cbpa.2009.09.029
Tung C-H, Lin Y, Moon WK, Weissleder R (2002) A receptor-targeted near-infrared fluorescence probe for in vivo tumor imaging. ChemBioChem 3(8):784–786. doi:10.1002/1439-7633(20020802)3:8<784:AID-CBIC784>3.0.CO;2-X
Zhang Z, Achilefu S (2005) Design, synthesis and evaluation of near-infrared fluorescent pH indicators in a physiologically relevant range. Chem Commun 47:5887–5889. doi:10.1039/B512315A
Weissleder R, Tung CH, Mahmood U, Bogdanov A Jr (1999) In vivo imaging of tumors with protease-activated near-infrared fluorescent probes. Nat Biotechnol 17(4):375–378. doi:10.1038/7933
Messerli SM, Prabhakar S, Tang Y, Shah K, Cortes ML, Murthy V, Weissleder R, Breakefield XO, Tung CH (2004) A novel method for imaging apoptosis using a caspase-1 near-infrared fluorescent probe. Neoplasia 6(2):95–105
Dadgar S, Ramjan Z, Floriano WB (2013) Paclitaxel Is an inhibitor and its boron dipyrromethene derivative is a fluorescent recognition agent for botulinum neurotoxin subtype A. J Med Chem 56(7):2791–2803
Shah F, Mukherjee P, Gut J, Legac J, Rosenthal PJ, Tekwani BL, Avery MA (2011) Identification of novel malarial cysteine protease inhibitors using structure-based virtual screening of a focused cysteine protease inhibitor library. J Chem Inf Model 51(4):852–864. doi:10.1021/ci200029y
Naylor E, Arredouani A, Vasudevan SR, Lewis AM, Parkesh R, Mizote A, Rosen D, Thomas JM, Izumi M, Ganesan A (2009) Identification of a chemical probe for NAADP by virtual screening. Nat Chem Biol 5(4):220–226
Doman TN, McGovern SL, Witherbee BJ, Kasten TP, Kurumbail R, Stallings WC, Connolly DT, Shoichet BK (2002) Molecular docking and high-throughput screening for novel inhibitors of protein tyrosine phosphatase-1B. J Med Chem 45(11):2213–2221. doi:10.1021/jm010548w
Kolb P, Rosenbaum DM, Irwin JJ, Fung JJ, Kobilka BK, Shoichet BK (2009) Structure-based discovery of β2-adrenergic receptor ligands. Proc Natl Acad Sci 106(16):6843–6848. doi:10.1073/pnas.0812657106
De Graaf C, Kooistra AJ, Vischer HF, Katritch V, Kuijer M, Shiroishi M, Iwata S, Shimamura T, Stevens RC, De Esch IJP, Leurs R (2011) Crystal structure-based virtual screening for fragment-like ligands of the human histamine H 1 receptor. J Med Chem 54(23):8195–8206
Floriano WB, Vaidehi N, Zamanakos G, Goddard WA (2004) HierVLS hierarchical docking protocol for virtual ligand screening of large-molecule databases. J Med Chem 47(1):56–71
Dadgar S, Chowdhury M, Phenix C, Floriano WB (2014) Systematic discovery of novel fluorescent molecular probes targeting multiple non-orthosteric spatially distinct sites in BoNTA (in preparation)
Kitchen DB, Decornez H, Furr JR, Bajorath J (2004) Docking and scoring in virtual screening for drug discovery: methods and applications. Nat Rev Drug Discov 3(11):935–949
Meng X-Y, Zhang H-X, Mezei M, Cui M (2011) Molecular docking: a powerful approach for structure-based drug discovery. Curr Comput Aided Drug Des 7(2):146
Scheffner M, Huibregtse JM, Vierstra RD, Howley PM (1993) The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell 75(3):495–505. doi:10.1016/0092-8674(93)90384-3
Huibregtse JM, Scheffner M, Howley PM (1991) A cellular protein mediates association of p53 with the E6 oncoprotein of human papillomavirus types 16 or 18. EMBO J 10(13):4129
Scheffner M, Werness BA, Huibregtse JM, Levine AJ, Howley PM (1990) The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell 63(6):1129–1136. doi:10.1016/0092-8674(90)90409-8
Sobel J, Tucker N, Sulka A, McLaughlin J, Maslanka S (2004) Foodborne botulism in the United States, 1990–2000. Emerg Infect Dis 10(9):1606
Cai S, Singh BR, Sharma S (2007) Botulism diagnostics: from clinical symptoms to in vitro assays. Crit Rev Microbiol 33(2):109–125
Arnon SS, Schechter R, Inglesby TV, Henderson DA, Bartlett JG, Ascher MS, Eitzen E, Fine AD, Hauer J, Layton M (2001) Botulinum toxin as a biological weapon. JAMA 285(8):1059–1070
Jankovic J (2004) Botulinum toxin in clinical practice. J Neurol Neurosurg Psychiatry 75(7):951–957
Münchau A, Bhatia K (2000) Regular review: uses of botulinum toxin injection in medicine today. Br Med J 320(7228):161
Bolton E, Wang Y, Thiessen PA, Bryant SH (2008) PubChem: integrated platform of small molecules and biological activities. In: Wheeler RA, Spellmeyer DC (eds) Annual reports in computational chemistry, vol 4. American Chemical Society, Washington, DC
Lipinski CA (2000) Drug-like properties and the causes of poor solubility and poor permeability. J Pharmacol Toxicol Methods 44(1):235–249. doi:10.1016/s1056-8719(00)00107-6
Molecular Operating Environment (MOE) (2010) 2010.10 edn. Chemical Computing Group, Inc., Montreal, Quebec, Canada
Gasteiger J, Marsili M (1980) Iterative partial equalization of orbital electronegativity—a rapid access to atomic charges. Tetrahedron 36:3219–3228. doi:10.1016/0040-4020(80)80168-2
Halgren TA (1996) Merck molecular force field. I. Basis, form, scope, parameterization, and performance of MMFF94. J Comput Chem 17(5–6):490–519
Mujumdar RB, Ernst L, Mujumdar SR, Lewis CJ, Waggoner AS (1993) Cyanine dye labeling reagents: sulfoindocyanine succinimidyl esters. Bioconjug Chem 4:105–111
Southwick P, Ernst L, Tauriello E (1990) Cyanine dye labeling reagents—carboxymethylindocyanine succinimidyl esters. Cytometry 430:418–430
Brady GP, Stouten PF (2000) Fast prediction and visualization of protein binding pockets with PASS. J Comput Aided Mol Des 14:383–401
PRISM (2013) 6.UPDATE for Windows* edn. GraphPad Software, La Jolla, California, USA
Program ML (2007) MLSMR compounds. https://mli.nih.gov/mli/compound-repository/mlsmr-compounds/. Accessed June 2013 2013
PubChem N (2011) PubChem3D release notes. http://pubchem.ncbi.nlm.nih.gov/release3d.html. Accessed June 2013
Halgren T (1996) Merck molecular force field. I. Basis, form, scope, parameterization, and performance of MMFF94. J Comput Chem 17:490–519
Carta G, Knox AJ, Lloyd DG (2007) Unbiasing scoring functions: a new normalization and rescoring strategy. J Chem Inf Model 47(4):1564–1571. doi:10.1021/ci600471m
Konstantinou-Kirtay C, Mitchell JB, Lumley JA (2007) Scoring functions and enrichment: a case study on Hsp90. BMC Bioinform 8:27. doi:10.1186/1471-2105-8-27
Jacobsson M, Karlen A (2006) Ligand bias of scoring functions in structure-based virtual screening. J Chem Inf Model 46(3):1334–1343. doi:10.1021/ci050407t
Pan Y, Huang N, Cho S, MacKerell AD Jr (2003) Consideration of molecular weight during compound selection in virtual target-based database screening. J Chem Inf Comput Sci 43(1):267–272. doi:10.1021/ci020055f
Allen M, Reeves J, Mellor G (2000) High throughput fluorescence polarization: a homogeneous alternative to radioligand binding for cell surface receptors. J Biomol Screen 5(2):63–69. doi:10.1177/108705710000500202
Jolley ME (1996) Fluorescence polarization assays for the detection of proteases and their inhibitors. J Biomol Screen 1(1):33–38
Checovich WJ, Bolger RE, Burke T (1995) Fluorescence polarization—a new tool for cell and molecular biology. Nature 375(6528):254–256
Warren GL, Andrews CW, Capelli A-M, Clarke B, LaLonde J, Lambert MH, Lindvall M, Nevins N, Semus SF, Senger S, Tedesco G, Wall ID, Woolven JM, Peishoff CE, Head MS (2005) A critical assessment of docking programs and scoring functions. J Med Chem 49(20):5912–5931. doi:10.1021/jm050362n
Buschmann V, Weston KD, Sauer M (2003) Spectroscopic study and evaluation of red-absorbing fluorescent dyes. Bioconjug Chem 14:195–204. doi:10.1021/bc025600x
Acknowledgments
This research was supported with funds from the Thunder Bay Regional Research Institute, the Natural Sciences and Engineering Research Council of Canada (NSERC), and the RBC Royal Bank’s Dr. Mark Poznansky Mentorship Development Award. High-throughput computational work in this project was made possible with resources provided through SHARCNET (www.sharcnet.ca) and Lakehead University’s High Performance Computing Centre (LUHPCC). The authors wish to acknowledge Darryl Willick for his help with data transfer and manipulation.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Kamstra, R.L., Dadgar, S., Wigg, J. et al. Creating and virtually screening databases of fluorescently-labelled compounds for the discovery of target-specific molecular probes. J Comput Aided Mol Des 28, 1129–1142 (2014). https://doi.org/10.1007/s10822-014-9789-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10822-014-9789-0