Abstract
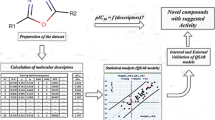
Improving performance of scoring functions for drug docking simulations is a challenging task in the modern discovery pipeline. Among various ways to enhance the efficiency of scoring function, tuning of energetic component approach is an attractive option that provides better predictions. Herein we present the first development of rapid and simple tuning models for predicting and scoring inhibitory activity of investigated ligands docked into catalytic core domain structures of HIV-1 integrase (IN) enzyme. We developed the models using all energetic terms obtained from flexible ligand-rigid receptor dockings by AutoDock4, followed by a data analysis using either partial least squares (PLS) or self-organizing maps (SOMs). The models were established using 66 and 64 ligands of mercaptobenzenesulfonamides for the PLS-based and the SOMs-based inhibitory activity predictions, respectively. The models were then evaluated for their predictability quality using closely related test compounds, as well as five different unrelated inhibitor test sets. Weighting constants for each energy term were also optimized, thus customizing the scoring function for this specific target protein. Root-mean-square error (RMSE) values between the predicted and the experimental inhibitory activities were determined to be <1 (i.e. within a magnitude of a single log scale of actual IC50 values). Hence, we propose that, as a pre-functional assay screening step, AutoDock4 docking in combination with these subsequent rapid weighted energy tuning methods via PLS and SOMs analyses is a viable approach to predict the potential inhibitory activity and to discriminate among small drug-like molecules to target a specific protein of interest.
Graphical Abstract








Similar content being viewed by others
References
Lahana R (1999) How many leads from HTS? Drug Discov Today 4:447–448
Lobanov V (2004) Using artificial neural networks to drive virtual screening of combinatorial libraries. Drug Discov Today Biosilico 2:149–156
Ou-Yang SS, Lu JY, Kong XQ, Liang ZJ, Luo C, Jiang H (2012) Computational drug discovery. Acta Pharmacol Sin 33:1131–1140
Ooms F (2000) Molecular modeling and computer aided drug design. Examples of their applications in medicinal chemistry. Curr Med Chem 7:141–158
Song CM, Lim SJ, Tong JC (2009) Recent advances in computer-aided drug design. Brief Bioinform 10:579–591
Meng XY, Zhang HX, Mezei M, Cui M (2011) Molecular docking: a powerful approach for structure-based drug discovery. Curr Comput Aided Drug Des 7:146–157
Waszkowycz B, Clark DE, Gancia E (2011) Outstanding challenges in protein–ligand docking and structure-based virtual screening. WIREs Comput Mol Sci 1:229–259
Renner S, Derksen S, Radestock S, Mörchen F (2008) Maximum common binding modes (MCBM): consensus docking scoring using multiple ligand information and interaction fingerprints. J Chem Inf Model 48:319–332
Coupez B, Lewis RA (2006) Docking and scoring—theoretically easy, practically impossible? Curr Med Chem 13:2995–3003
Zeng FQ, Peng SM, Li L, Mu LB, Zhang ZH, Zhang ZY, Huang N (2013) Structure-based identification of drug-like inhibitors of p300 histone acetyltransferase. Yao Xue Xue Bao 48:700–708
Vidler LR, Filippakopoulos P, Fedorov O, Picaud S, Martin S, Tomsett M, Woodward H, Brown N, Knapp S, Hoelder S (2013) Discovery of novel small-molecule inhibitors of BRD4 using structure-based virtual screening. J Med Chem 56:8073–8088
Sorna V, Theisen ER, Stephens B, Warner SL, Bearss DJ, Vankayalapati H, Sharma S (2013) High-throughput virtual screening identifies novel N′-(1-phenylethylidene)-benzohydrazides as potent, specific, and reversible LSD1 inhibitors. J Med Chem 56:9496–9508
Azam SS, Abbasi SW, Tahir S (2014) Investigation of novel chemical inhibitors of human lysosomal acid lipase: virtual screening and molecular docking studies. Comb Chem High Throughput Screen 17:473–482
Bandaru S, Marri VK, Kasera P, Kovuri P, Girdhar A, Mittal DR, Ikram S, Gv R, Nayarisseri A (2014) Structure based virtual screening of ligands to identify cysteinyl leukotriene receptor 1 antagonist. Bioinformation 10:652–657
Doig P, Boriack-Sjodin PA, Dumas J, Hu J, Itoh K, Johnson K, Kazmirski S, Kinoshita T, Kuroda S, Sato TO, Sugimoto K, Tohyama K, Aoi H, Wakamatsu K, Wang H (2014) Rational design of inhibitors of the bacterial cell wall synthetic enzyme glmu using virtual screening and lead-hopping. Bioorg Med Chem 22:6256–6269
Yang C, Chen S, Zhou M, Li Y, Zhang Z, Liu Z, Ba Q, Li J, Wang H, Yan X, Ma D, Wang R (2014) Development of 3-phenyl-N-(2-(3-phenylureido)ethyl)-thiophene-2-sulfonamide compounds as inhibitors of antiapoptotic Bcl-2 family proteins. Chem Med Chem 9:1436–1452
Zheng GH, Shen JJ, Zhan YC, Yi H, Xue ST, Wang Z, Ji XY, Li ZR (2014) Design, synthesis and in vitro and in vivo antitumour activity of 3-benzylideneindolin-2-one derivatives, a novel class of small-molecule inhibitors of the MDM2-p53 interaction. Eur J Med Chem 81:277–288
Gohlke H, Hendlich M, Klebe G (2000) Knowledge-based scoring function to predict protein–ligand interactions. J Mol Biol 295:337–356
Gohlke H, Klebe G (2002) Approaches to the description and prediction of the binding affinity of small-molecule ligands to macromolecular receptors. Angew Chem Int Ed 41:2644–2676
Kroemer RT (2007) Structure-based drug design: docking and scoring. Curr Protein Pept Sci 8:312–328
Williams DH, Maguire AJ, Tsuzuki W, Westwell MS (1998) An analysis of the origins of a cooperative binding energy of dimerization. Science 280:711–714
Huey R, Morris GM, Olson AJ, Goodsell DS (2007) A semiempirical free energy force field with charge-based desolvation. J Comput Chem 28:1145–1152
Wang T, Wade RC (2001) Comparative binding energy (COMBINE) analysis of influenza neuraminidase-inhibitor complexes. J Med Chem 44:961–971
Murcia M, Ortiz AR (2004) Virtual screening with flexible docking and COMBINE-based models. Application to a series of factor Xa inhibitors. J Med Chem 47:805–820
Charifson PS, Corkery JJ, Murcko MA, Walters WP (1999) Consensus scoring: a method for obtaining improved hit rates from docking databases of three-dimensional structures into proteins. J Med Chem 42:5100–5109
Clark RD, Strizhev A, Leonard JM, Blake JF, Matthew JB (2002) Consensus scoring for ligand/protein interactions. J Mol Graph Model 20:281–295
Schultes S, Kooistra AJ, Vischer HF, Nijmeijer S, Haaksma EE, Leurs R, de Esch IJ, de Graaf C (2015) Combinatorial consensus scoring for ligand-based virtual fragment screening: a comparative case study for serotonin 5-HT3A, histamine H1, and histamine H4 receptors. J Chem Inf Model 55:1030–1044
Kuo CL, Assefa H, Kamath S, Brzozowski Z, Slawinski J, Saczewski F, Buolamwini JK, Neamati N (2004) Application of CoMFA and CoMSIA 3D-QSAR and docking studies in optimization of mercaptobenzenesulfonamides as HIV-1 integrase inhibitors. J Med Chem 47:385–399
Brzozowski Z, Saczewski F, Sanchez T, Kuo CL, Gdaniec M, Neamati N (2004) Synthesis, antiviral, and anti-HIV-1 integrase activities of 3-aroyl-1,1-dioxo-1,4,2-benzodithiazines. Bioorg Med Chem 12:3663–3672
Brzozowski Z, Saczewski F, Sławiński J, Sanchez T, Neamati N (2009) Synthesis and anti-HIV-1 integrase activities of 3-aroyl-2,3-dihydro-1,1-dioxo-1,4,2-benzodithiazines. Eur J Med Chem 44:190–196
Lin Z, Neamati N, Zhao H, Kiryu Y, Turpin JA, Aberham C, Strebel K, Kohn K, Witvrouw M, Pannecouque C, Debyser Z, De Clercq E, Rice WG, Pommier Y, Burke TR (1999) Chicoric acid analogues as HIV-1 integrase inhibitors. J Med Chem 42:1401–1414
Sechi M, Derudas M, Dallocchio R, Dessì A, Bacchi A, Sannia L, Carta F, Palomba M, Ragab O, Chan C, Shoemaker R, Sei S, Dayam R, Neamati N (2004) Design and synthesis of novel indole β-diketo acid derivatives as HIV-1 integrase inhibitors. J Med Chem 47:5298–5310
Korolev SP, Kondrashina OV, Druzhilovsky DS, Starosotnikov AM, Dutov MD, Bastrakov MA, Dalinger IL, Filimonov DA, Shevelev SA, Poroikov VV, Agapkina YY, Gottikh MB (2013) Structural-functional analysis of 2,1,3-benzoxadiazoles and their N-oxides as HIV-1 integrase inhibitors. Acta Naturae 5:63–72
Goldgur Y, Craigie R, Cohen GH, Fujiwara T, Yoshinaga T, Fujishita T, Sugimoto H, Endo T, Murai H, Davies DR (1999) Structure of the HIV-1 integrase catalytic domain complexed with an inhibitor: a platform for antiviral drug design. Proc Natl Acad Sci USA 96:13040–13043
Maignan S, Guilloteau JP, Zhou-Liu Q, Clément-Mella C, Mikol V (1998) Crystal structures of the catalytic domain of HIV-1 integrase free and complexed with its metal cofactor: high level of similarity of the active site with other viral integrases. J Mol Biol 282:359–368
Goldgur Y, Dyda F, Hickman AB, Jenkins TM, Craigie R, Davies DR (1998) Three new structures of the core domain of HIV-1 integrase: an active site that binds magnesium. Proc Natl Acad Sci USA 95:9150–9154
Dill KA (1997) Additivity principles in biochemistry. J Biol Chem 272:701–704
Klebe G (2015) Applying thermodynamic profiling in lead finding and optimization. Nat Rev Drug Discov 14:95–110
Helland IS (1988) On the structure of partial least squares regression. Commun Stat Simul Comput 17:581–607
Wold S, Sjostrom M, Eriksson L (2001) PLS-regression: a basic tool of chemometrics. Chemometr Intell Lab Syst 58:109–130
Brereton RG (2012) Self organising maps for visualising and modelling. Chem Cent J 6:S1
Kittiwachana S, Wangkarn S, Grudpan K, Brereton RG (2013) Prediction of liquid chromatographic retention behavior based on quantum chemical parameters using supervised self organizing maps. Talanta 106:229–236
Ultsch A (1993) In: Information and classification (ed) Self-organizing neural networks for visualisation and classification. Springer, Berlin
Di Santo R (2011) Diketo acids derivatives as dual inhibitors of human immunodeficiency virus type 1 integrase and the reverse transcriptase RNase H domain. Curr Med Chem 18:3335–3342
Huang M, Grant GH, Richards WG (2011) Binding modes of diketo-acid inhibitors of HIV-1 integrase: a comparative molecular dynamics simulation study. J Mol Graph Model 29:956–964
Johns BA, Svolto AC (2008) Advances in two-metal chelation inhibitors of HIV integrase. Expert Opin 18:1225–1237
Bacchi A, Carcelli M, Compari C, Fisicaro E, Pala N, Rispoli G, Rogolino D, Sanchez TW, Sechi M, Sinisi V, Neamati N (2011) Investigating the role of metal chelation in HIV-1 integrase strand transfer inhibitors. J Med Chem 54:8407–8420
Bacchi A, Carcelli M, Compari C, Fisicaro E, Pala N, Rispoli G, Rogolino D, Sanchez TW, Sechi M, Neamati N (2011) HIV-1 IN strand transfer chelating inhibitors: a focus on metal binding. Mol Pharm 8:507–519
Agrawal A, DeSoto J, Fullagar JL, Maddali K, Rostami S, Richman DD, Pommier Y, Cohen SM (2012) Probing chelation motifs in HIV integrase inhibitors. Proc Natl Acad Sci USA 109:2251–2256
Ribeiro AJ, Ramos MJ, Fernandes PA (2012) The catalytic mechanism of HIV-1 integrase for DNA 3′-end processing established by QM/MM calculations. J Am Chem Soc 134:13436–13447
Murray CW, Baxter CA, Frenkel AD (1999) The sensitivity of the results of molecular docking to induced fit effects: application to thrombin, thermolysin and neuraminidase. J Comput Aided Mol Des 13:547–562
Perez C, Ortiz AR (2001) Evaluation of docking functions for protein-ligand docking. J Med Chem 44:3768–3785
Ferrara P, Gohlke H, Price DJ, Klebe G, Brooks CL (2004) Assessing scoring functions for protein–ligand interactions. J Med Chem 47:3032–3047
Guo L, Yan Z, Zheng X, Hu L, Yang Y, Wang J (2014) A comparison of various optimization algorithms of protein–ligand docking programs by fitness accuracy. J Mol Model 20:2251
Boulesteix AL, Strimmer K (2007) Partial least squares: a versatile tool for the analysis of high-dimensional genomic data. Brief Bioinform 8:32–44
Haritopoulos M, Yin H, Allinson NM (2002) Image denoising using self-organizing map-based nonlinear independent component analysis. Neural Netw 15:1085–1098
Xiao YD, Clauset A, Harris R, Bayram E, Santago P, Schmitt JD (2005) Supervised self-organizing maps in drug discovery. 1. Robust behavior with overdetermined data sets. J Chem Inf Model 45:1749–1758
Xiao YD, Harris R, Bayram E, Ii PS, Schmitt JD (2006) Supervised self-organizing maps in drug discovery. 2. Improvements in descriptor selection and model validation. J Chem Inf Model 46:137–144
Acknowledgments
This work was funded by the Junior Science Talent Project (JSTP) from the National Science and Technology Development Agency (NSTDA) of Thailand, and the Research Fund for DPST Graduate with First Placement (Grant: 037/2555). N.S. was supported by the Grant for New Researcher (Grant: TRG5880271) from the Thailand Research Fund (TRF); Chiang Mai University Young Faculty Research Grant; the National Research Council of Thailand (Grant: 2556NRCT51390 and 2558NRCT350269); the National Research University Project under Thailand’s Office of the Higher Education Commission; P.T. was supported by the JSTP-NSTDA research grant for graduate study (Grant: JSTP-06-55-35E). We would like to thank the Computer Technology Center of NECTEC for the SYBYL software, the Drug Discovery Research Laboratory and the Department of Chemistry, Faculty of Science, Chiang Mai University for all research facilities. We also thank Prof. Richard Deming (CSU Fullerton) for helpful suggestions.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Thangsunan, P., Kittiwachana, S., Meepowpan, P. et al. Rapid activity prediction of HIV-1 integrase inhibitors: harnessing docking energetic components for empirical scoring by chemometric and artificial neural network approaches. J Comput Aided Mol Des 30, 471–488 (2016). https://doi.org/10.1007/s10822-016-9917-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10822-016-9917-0