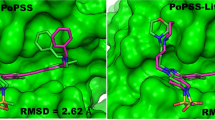
Abstract
Ligand-based virtual screening has proven to be a viable technology during the search for new lead structures in drug discovery. Despite the rapidly increasing number of published methods, meaningful shape matching as well as ligand and target flexibility still remain open challenges. In this work, we analyze the influence of knowledge-based sterical constraints on the performance of the recently published ligand-based virtual screening method mRAISE. We introduce the concept of partial shape matching enabling a more differentiated view on chemical structure. The new method is integrated into the LBVS tool mRAISE providing multiple options for such constraints. The applied constraints can either be derived automatically from a protein–ligand complex structure or by manual selection of ligand atoms. In this way, the descriptor directly encodes the fit of a ligand into the binding site. Furthermore, the conservation of close contacts between the binding site surface and the query ligand can be enforced. We validated our new method on the DUD and DUD-E datasets. Although the statistical performance remains on the same level, detailed analysis reveal that for certain and especially very flexible targets a significant improvement can be achieved. This is further highlighted looking at the quality of calculated molecular alignments using the recently introduced mRAISE dataset. The new partial shape constraints improved the overall quality of molecular alignments especially for difficult targets with highly flexible or different sized molecules. The software tool mRAISE is freely available on Linux operating systems for evaluation purposes and academic use (see http://www.zbh.uni-hamburg.de/raise).








Similar content being viewed by others
References
Lavecchia A, Di Giovanni C (2013) Virtual screening strategies in drug discovery: a critical review. Curr Med Chem 20(23):2839–2860
Schomburg KT, Bietz S, Briem H, Henzler AM, Urbaczek S, Rarey M (2014) Facing the challenges of structure-based target prediction by inverse virtual screening. J Chem Inf Model 54:1676–1686
Henzler AM, Urbaczek S, Hilbig M, Rarey M (2014) An integrated approach to knowledge-driven structure-based virtual screening. J Comput Aided Mol Des 28:927–939
von Behren MM, Bietz S, Nittinger E, Rarey M (2016) mRAISE: an alternative algorithmic approach to ligand-based virtual screening. J Comput Aided Mol Des 30:583–594
Grant JA, Gallardo MA, Pickup BT (1996) A fast method of molecular shape comparison: a simple application of a gaussian description of molecular shape. J Comput Chem 17(14):1653–1666
Vainio MJ, Puranen JS, Johnson MS (2009) ShaEP: molecular overlay based on shape and electrostatic potential. J Chem Inf Model 49:492–502
Vaz de Lima LA, Nascimento AS (2013) MolShaCS: a free and open source tool for ligand similarity identification based on Gaussian descriptors. Eur J Med Chem 59:296–303
Roy A, Skolnick J (2015) LIGSIFT: an open-source tool for ligand structural alignment and virtual screening. Bioinformatics 31:539–544
Taminau J, Thijs G, De Winter H (2008) Pharao: pharmacophore alignment and optimization. J Mol Graph Model 27:161–169
Nittinger E, Schneider N, Lange G, Rarey M (2015) Evidence of water molecules-a statistical evaluation of water molecules based on electron density. J Chem Inf Model 55:771–783
von Behren MM, Volkamer A, Henzler AM, Schomburg KT, Urbaczek S, Rarey M (2013) Fast protein binding site comparison via an index-based screening technology. J Chem Inf Model 53(2):411–422
Kitchen DB, Decornez H, Furr JR, Bajorath J (2004) Docking and scoring in virtual screening for drug discovery: methods and applications. Nat Rev Drug Discov 3:935–949
Wei DQ, Zhang R, Du QS, Gao WN, Li Y, Gao H, Wang SQ, Zhang X, Li AX, Sirois S, Chou KC (2006) Anti-SARS drug screening by molecular docking. Amino Acids 31:73–80
Barreiro G, Guimaraes CR, Tubert-Brohman I, Lyons TM, Tirado-Rives J, Jorgensen WL (2007) Search for non-nucleoside inhibitors of HIV-1 reverse transcriptase using chemical similarity, molecular docking, and MM-GB/SA scoring. J Chem Inf Model 47(6):2416–2428
Tikhonova IG, Sum CS, Neumann S, Engel S, Raaka BM, Costanzi S, Gershengorn MC (2008) Discovery of novel agonists and antagonists of the free fatty acid receptor 1 (FFAR1) using virtual screening. J Med Chem 51:625–633
Lin TW, Melgar MM, Kurth D, Swamidass SJ, Purdon J, Tseng T, Gago G, Baldi P, Gramajo H, Tsai SC (2006) Structure-based inhibitor design of AccD5, an essential acyl-CoA carboxylase carboxyltransferase domain of Mycobacterium tuberculosis. Proc Natl Acad Sci USA 103:3072–3077
Vidal D, Thormann M, Pons M (2006) A novel search engine for virtual screening of very large databases. J Chem Inf Model 46(2):836–843
Mestres J, Knegtel RM (2000) Similarity versus docking in 3d virtual screening. Perspect Drug Discov Des 20(1):191–207
Huang N, Shoichet Brian K, Irwin JJ (2006) Benchmarking sets for molecular docking. J Med Chem 49(23):6789–6801
Mysinger MM, Carchia M, Irwin JJ, Shoichet BK (2012) Directory of useful decoys, enhanced (DUD-E): better ligands and decoys for better benchmarking. J. Med. Chem. 55:6582–6594
Wu K (2005) Fastbit: an efficient indexing technology for accelerating data-intensive science. J Phys 16(1):556
Bietz S, Rarey M (2016) SIENA: efficient compilation of selective protein binding site ensembles. J Chem Inf Model 56:248–259
Hilbig M, Rarey M (2015) MONA 2: a light cheminformatics platform for interactive compound library processing. J Chem Inf Model 55:2071–2078
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
von Behren, M.M., Rarey, M. Ligand-based virtual screening under partial shape constraints. J Comput Aided Mol Des 31, 335–347 (2017). https://doi.org/10.1007/s10822-017-0011-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10822-017-0011-z