Abstract
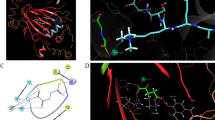
The protein lysine methyltransferase G9a, which controls gene expression by epigenetic regulation of H3K9 methylation, is related to various human diseases, including cancer, drug addiction, and mental retardation. In recent years, genetic, biological, and physiological evidence has established G9a inhibitors as potential chemotherapeutic agents for cancer treatment. In this study, we identified protoberberine alkaloid pseudodehydrocorydaline (CT13) as a novel G9a inhibitor, by structure-based virtual screening of in-house library containing natural product compounds. The activity of CT13 was determined by biophysical analyses involving MALDI-TOF mass spectrometry and western blot analysis. CT13 showed selective inhibitory activity against G9a and suppressed the level of H3K9me2 in MCF7 human breast cancer cells. Molecular docking analysis suggested the binding mode of CT13 which occupies the binding site of histone H3 substrate. CT13 provides a novel scaffold for further development of analogous synthetic G9a inhibitors.
Graphical abstract







Similar content being viewed by others
Abbreviations
- CDP/cut:
-
CCAAT displacement protein/cut homolog
- EHMT1:
-
Euchromatic histone lysine methyltransferase 1
- EHMT2:
-
Euchromatic histone lysine methyltransferase 2
- Gfi1:
-
Growth factor independent 1 transcription repressor
- GLP:
-
G9a like protein
- H3K9:
-
Histone H3 lysine 9
- H3K9Me2:
-
Histone H3 lysine 9 dimethylation
- HIV-1:
-
Human immunodeficiency virus 1
- PKMTs:
-
Protein lysine methyltransferases
- PRMTs:
-
Protein arginine methyltransferases
- PTMs:
-
Post-translational modifications
- SAH:
-
S-5′-adenosyl-l-homocysteine
- SAM:
-
S-5′-adenosyl-l-methionine
- UHRF1:
-
Ubiquitin-like with PHD and ring finger domains 1
- WIZ:
-
Widely interspaced zinc finger motifs protein
References
Rea S, Eisenhaber F, O’Carroll D, Strahl BD, Sun ZW, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD, Jenuwein T (2000) Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 406:593–599. https://doi.org/10.1038/35020506
Fog CK, Jensen KT, Lund AH (2007) Chromatin-modifying proteins in cancer. Apmis 115(10):1060–1089. https://doi.org/10.1111/j.1600-0463.2007.apm_776.xml.x
Copeland RA, Solomon ME, Richon VM (2009) Protein methyltransferases as a target class for drug discovery. Nat Rev Drug Discov 8(9):724–732. https://doi.org/10.1038/nrd2974
Spannhoff A, Hauser AT, Heinke R, Sippl W, Jung M (2009) The emerging therapeutic potential of histone methyltransferase and demethylase inhibitors. ChemMedChem 4(10):1568–1582. https://doi.org/10.1002/cmdc.200900301
Spannhoff A, Sippl W, Jung M (2009) Cancer treatment of the future: inhibitors of histone methyltransferases. Int J Biochem Cell Biol 41(1):4–11. https://doi.org/10.1016/j.biocel.2008.07.024
Li Y, Reddy MA, Miao F, Shanmugam N, Yee JK, Hawkins D, Ren B, Natarajan R (2008) Role of the histone H3 lysine 4 methyltransferase, SET7/9, in the regulation of NF-kappaB-dependent inflammatory genes. Relevance to diabetes and inflammation. J Biol Chem 283(39):26771–26781. https://doi.org/10.1074/jbc.M802800200
Maze I, Covington HE, Dietz DM, LaPlant Q, Renthal W, Russo SJ, Mechanic M, Mouzon E, Neve RL, Haggarty SJ, Ren Y, Sampath SC, Hurd YL, Greengard P, Tarakhovsky A, Schaefer A, Nestler EJ (2010) Essential role of the histone methyltransferase G9a in cocaine-induced plasticity. Science 327(5962):213–216. https://doi.org/10.1126/science.1179438
Tachibana M, Sugimoto K, Fukushima T, Shinkai Y (2001) Set domain-containing protein, G9a, is a novel lysine-preferring mammalian histone methyltransferase with hyperactivity and specific selectivity to lysines 9 and 27 of histone H3. J Biol Chem 276(27):25309–25317. https://doi.org/10.1074/jbc.M101914200
Tachibana M, Ueda J, Fukuda M, Takeda N, Ohta T, Iwanari H, Sakihama T, Kodama T, Hamakubo T, Shinkai Y (2005) Histone methyltransferases G9a and GLP form heteromeric complexes and are both crucial for methylation of euchromatin at H3-K9. Genes Dev 19(7):815–826. https://doi.org/10.1101/gad.1284005
Nishio H, Walsh MJ (2004) CCAAT displacement protein cut homolog recruits G9a histone lysine methyltransferase to repress transcription. Proc Natl Acad Sci USA 101(31):11257–11262
Duan Z, Zarebski A, Montoya-Durango D, Grimes HL, Horwitz M (2005) Gfi1 coordinates epigenetic repression of p21Cip/WAF1 by recruitment of histone lysine methyltransferase G9a and histone deacetylase 1. Mol Cell Biol 25(23):10338–10351. https://doi.org/10.1128/MCB.25.23.10338-10351.2005
Ueda J, Tachibana M, Ikura T, Shinkai Y (2006) Zinc finger protein Wiz links G9a/GLP histone methyltransferases to the co-repressor molecule CtBP. J Biol Chem 281(29):20120–20128. https://doi.org/10.1074/jbc.M603087200
Kim JK, Esteve PO, Jacobsen SE, Pradhan S (2009) UHRF1 binds G9a and participates in p21 transcriptional regulation in mammalian cells. Nucleic Acids Res 37(2):493–505. https://doi.org/10.1093/nar/gkn961
Kondo Y, Shen L, Suzuki S, Kurokawa T, Masuko K, Tanaka Y, Kato H, Mizuno Y, Yokoe M, Sugauchi F, Hirashima N, Orito E, Osada H, Ueda R, Guo Y, Chen X, Issa JP, Sekido Y (2007) Alterations of DNA methylation and histone modifications contribute to gene silencing in hepatocellular carcinomas. Hepatol Res 37(11):974–983. https://doi.org/10.1111/j.1872-034X.2007.00141.x
Huang J, Dorsey J, Chuikov S, Perez-Burgos L, Zhang X, Jenuwein T, Reinberg D, Berger SL (2010) G9a and Glp methylate lysine 373 in the tumor suppressor p53. J Biol Chem 285(13):9636–9641. https://doi.org/10.1074/jbc.M109.062588
Watanabe H, Soejima K, Yasuda H, Kawada I, Nakachi I, Yoda S, Naoki K, Ishizaka A (2008) Deregulation of histone lysine methyltransferases contributes to oncogenic transformation of human bronchoepithelial cells. Cancer Cell Int 8:15. https://doi.org/10.1186/1475-2867-8-15
Yang Q, Lu Z, Singh D, Raj JU (2012) BIX-01294 treatment blocks cell proliferation, migration and contractility in ovine foetal pulmonary arterial smooth muscle cells. Cell Prolif 45(4):335–344. https://doi.org/10.1111/j.1365-2184.2012.00828.x
Yuan Y, Wang Q, Paulk J, Kubicek S, Kemp MM, Adams DJ, Shamji AF, Wagner BK, Schreiber SL (2012) A small-molecule probe of the histone methyltransferase G9a induces cellular senescence in pancreatic adenocarcinoma. ACS Chem Biol 7(7):1152–1157. https://doi.org/10.1021/cb300139y
Kim Y, Kim YS, Kim DE, Lee JS, Song JH, Kim HG, Cho DH, Jeong SY, Jin DH, Jang SJ, Seol HS, Suh YA, Lee SJ, Kim CS, Koh JY, Hwang JJ (2013) BIX-01294 induces autophagy-associated cell death via EHMT2/G9a dysfunction and intracellular reactive oxygen species production. Autophagy 9(12):2126–2139. https://doi.org/10.4161/auto.26308
Ke XX, Zhang D, Zhu S, Xia Q, Xiang Z, Cui H (2014) Inhibition of H3K9 methyltransferase G9a repressed cell proliferation and induced autophagy in neuroblastoma cells. PLoS ONE 9(9):e106962. https://doi.org/10.1371/journal.pone.0106962
Li KC, Hua KT, Lin YS, Su CY, Ko JY, Hsiao M, Kuo ML, Tan CT (2014) Inhibition of G9a induces DUSP4-dependent autophagic cell death in head and neck squamous cell carcinoma. Mol Cancer 13(1):172. https://doi.org/10.1186/1476-4598-13-172
Schaefer A, Sampath SC, Intrator A, Min A, Gertler TS, Surmeier DJ, Tarakhovsky A, Greengard P (2009) Control of cognition and adaptive behavior by the GLP/G9a epigenetic suppressor complex. Neuron 64(5):678–691. https://doi.org/10.1016/j.neuron.2009.11.019
Imai K, Togami H, Okamoto T (2010) Involvement of histone H3 lysine 9 (H3K9) methyltransferase G9a in the maintenance of HIV-1 latency and its reactivation by BIX01294. J Biol Chem 285(22):16538–16545. https://doi.org/10.1074/jbc.M110.103531
Kubicek S, O’Sullivan RJ, August EM, Hickey ER, Zhang Q, Teodoro ML, Rea S, Mechtler K, Kowalski JA, Homon CA, Kelly TA, Jenuwein T (2007) Reversal of H3K9me2 by a small-molecule inhibitor for the G9a histone methyltransferase. Mol Cell 25(3):473–481. https://doi.org/10.1016/j.molcel.2007.01.017
Vedadi M, Barsyte-Lovejoy D, Liu F, Rival-Gervier S, Allali-Hassani A, Labrie V, Wigle TJ, Dimaggio PA, Wasney GA, Siarheyeva A, Dong A, Tempel W, Wang SC, Chen X, Chau I, Mangano TJ, Huang XP, Simpson CD, Pattenden SG, Norris JL, Kireev DB, Tripathy A, Edwards A, Roth BL, Janzen WP, Garcia BA, Petronis A, Ellis J, Brown PJ, Frye SV, Arrowsmith CH, Jin J (2011) A chemical probe selectively inhibits G9a and GLP methyltransferase activity in cells. Nat Chem Biol 7(8):566–574. https://doi.org/10.1038/nchembio.599
Kondengaden SM, Luo LF, Huang K, Zhu M, Zang L, Bataba E, Wang R, Luo C, Wang B, Li KK, Wang PG (2016) Discovery of novel small molecule inhibitors of lysine methyltransferase G9a and their mechanism in leukemia cell lines. Eur J Med Chem 122:382–393. https://doi.org/10.1016/j.ejmech.2016.06.028
Greiner D, Bonaldi T, Eskeland R, Roemer E, Imhof A (2005) Identification of a specific inhibitor of the histone methyltransferase SU(VAR)3-9. Nat Chem Biol 1(3):143–145. https://doi.org/10.1038/nchembio721
Cherblanc FL, Chapman KL, Brown R, Fuchter MJ (2013) Chaetocin is a nonspecific inhibitor of histone lysine methyltransferases. Nat Chem Biol 9(3):136–137. https://doi.org/10.1038/nchembio.1187
Wang J, Chen L, Sinha SH, Liang Z, Chai H, Muniyan S, Chou YW, Yang C, Yan L, Feng Y, Li KK, Lin MF, Jiang H, Zheng YG, Luo C (2012) Pharmacophore-based virtual screening and biological evaluation of small molecule inhibitors for protein arginine methylation. J Med Chem 55(18):7978–7987. https://doi.org/10.1021/jm300521m
Medina-Franco JL, Yoo J (2013) Molecular modeling and virtual screening of DNA methyltransferase inhibitors. Curr Pharm Des 19(12):2138–2147
Xie Y, Zhou R, Lian F, Liu Y, Chen L, Shi Z, Zhang N, Zheng M, Shen B, Jiang H, Liang Z, Luo C (2014) Virtual screening and biological evaluation of novel small molecular inhibitors against protein arginine methyltransferase 1 (PRMT1). Org Biomol Chem 12(47):9665–9673. https://doi.org/10.1039/c4ob01591f
Meng F, Cheng S, Ding H, Liu S, Liu Y, Zhu K, Chen S, Lu J, Xie Y, Li L, Liu R, Shi Z, Zhou Y, Liu YC, Zheng M, Jiang H, Lu W, Liu H, Luo C (2015) Discovery and optimization of novel, selective histone methyltransferase SET7 inhibitors by pharmacophore- and docking-based virtual screening. J Med Chem 58(20):8166–8181. https://doi.org/10.1021/acs.jmedchem.5b01154
Chen S, Wang Y, Zhou W, Li S, Peng J, Shi Z, Hu J, Liu YC, Ding H, Lin Y, Li L, Cheng S, Liu J, Lu T, Jiang H, Liu B, Zheng M, Luo C (2014) Identifying novel selective non-nucleoside DNA methyltransferase 1 inhibitors through docking-based virtual screening. J Med Chem 57(21):9028–9041. https://doi.org/10.1021/jm501134e
C.L.P. SX, 2.1 ed. SYBYL molecular modeling software (2013), St. Louis, MO
Liu F, Chen X, Allali-Hassani A, Quinn AM, Wasney GA, Dong A, Barsyte D, Kozieradzki I, Senisterra G, Chau I, Siarheyeva A, Kireev DB, Jadhav A, Herold JM, Frye SV, Arrowsmith CH, Brown PJ, Simeonov A, Vedadi M, Jin J (2009) Discovery of a 2,4-diamino-7-aminoalkoxyquinazoline as a potent and selective inhibitor of histone lysine methyltransferase G9a. J Med Chem 52(24):7950–7953. https://doi.org/10.1021/jm901543m
Jain AN (2003) Surflex: fully automatic flexible molecular docking using a molecular similarity-based search engine. J Med Chem 46(4):499–511. https://doi.org/10.1021/jm020406h
Clark RD, Strizhev A, Leonard JM, Blake JF, Matthew JB (2002) Consensus scoring for ligand/protein interactions. J Mol Graph Model 20(4):281–295. https://doi.org/10.1016/s1093-3263(01)00125-5
Friesner RA, Banks JL, Murphy RB, Halgren TA, Klicic JJ, Mainz DT, Repasky MP, Knoll EH, Shelley M, Perry JK, Shaw DE, Francis P, Shenkin PS (2004) Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J Med Chem 47(7):1739–1749. https://doi.org/10.1021/jm0306430
Friesner RA, Murphy RB, Repasky MP, Frye LL, Greenwood JR, Halgren TA, Sanschagrin PC, Mainz DT (2006) Extra precision glide: docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J Med Chem 49(21):6177–6196. https://doi.org/10.1021/jm051256o
Jonathan BB, Georgina AH (2010) New substructure filters for removal of pan assay interference compounds (PAINS) from screening libraries and for their exclusion in bioassays. J Med Chem 29(11):1039–1045. https://doi.org/10.1021/jm901137j
Anastassiadis T, Deacon SW, Devarajan K, Ma H, Peterson JR (2011) Comprehensive assay of kinase catalytic activity reveals features of kinase inhibitor selectivity. Nat Biotechnol 29(11):1039–1045. https://doi.org/10.1038/nbt.2017
Kim KH, Piao CJ, Choi SU, Son MW, Lee KR (2010) New cytotoxic tetrahydroprotoberberine-aporphine dimeric and aporphine alkaloids from Corydalis turtschaninovii. Planta Med 76(15):1732–1738. https://doi.org/10.1055/s-0030-1249972
Iranshahy M, Quinn RJ, Iranshahi M (2014) Biologically active isoquinoline alkaloids with drug-like properties from the genus Corydalis. RSC Adv. https://doi.org/10.1039/c3ra47944g
Jayaram H, Hoelper D, Jain SU, Cantone N, Lundgren SM, Poy F, Allis CD, Cummings R, Bellon S, Lewis PW (2016) S-adenosyl methionine is necessary for inhibition of the methyltransferase G9a by the lysine 9 to methionine mutation on histone H3. Proc Natl Acad Sci USA 113(22):6182–6187. https://doi.org/10.1073/pnas.1605523113
Wu H, Min J, Lunin VV, Antoshenko T, Dombrovski L, Zeng H, Allali-Hassani A, Campagna-Slater V, Vedadi M, Arrowsmith CH, Plotnikov AN, Schapira M (2010) Structural biology of human H3K9 methyltransferases. PLoS ONE 5(1):e8570. https://doi.org/10.1371/journal.pone.0008570
Hung TM, Na M, Dat NT, Ngoc TM, Youn U, Kim HJ, Min BS, Lee J, Bae K (2008) Cholinesterase inhibitory and anti-amnesic activity of alkaloids from Corydalis turtschaninovii. J Ethnopharmacol 119(1):74–80. https://doi.org/10.1016/j.jep.2008.05.041
Kim JH, Ryu YB, Lee WS, Kim YH (2014) Neuraminidase inhibitory activities of quaternary isoquinoline alkaloids from Corydalis turtschaninovii rhizome. Bioorg Med Chem 22(21):6047–6052. https://doi.org/10.1016/j.bmc.2014.09.004
Funding
This research was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (NRF-2012R1A5A2A28671860).
Author information
Authors and Affiliations
Contributions
H-JP and JC conceived of this project and designed the experiments; JC performed the virtual screening, docking simulation and biochemical assays; XL contributed to western blot analysis; KRL and KJP contributed to the isolation and preparation of the natural products. All authors have approved the final version of this manuscript.
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Chen, J., Lin, X., Park, K.J. et al. Identification of protoberberine alkaloids as novel histone methyltransferase G9a inhibitors by structure-based virtual screening. J Comput Aided Mol Des 32, 917–928 (2018). https://doi.org/10.1007/s10822-018-0156-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10822-018-0156-4