Abstract
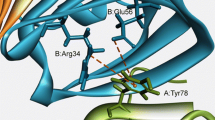
Anion binding CαNN motif is found in functionally important regions of protein structures. This motif based only on backbone atoms from three adjacent residues, recognizes free sulphate or phosphate ion as well as phosphate groups in nucleotides and in a variety of cofactors. The mode of anion recognition and microscopic picture of binding interaction remains unclear. Here we perform self-consistent quantum chemical calculations considering sulphate and phosphate bound CαNN motif fragments from crystal structures of functional proteins in order to figure out microscopic basis of anion recognition. Our calculations indicate that stability and preference of the anion in the motif depends on the sequence of the motif. The stabilization energy is larger in case of polar residue containing motif fragment. Nitrogen atom of the polar residue of motif mainly participates in the coordination at the lowest energy levels. Anion replacement decreases stabilization energy along with coordination between motif atoms and oxygen atoms of anion shifted to higher energies, suggesting preference of the motif residues to specific anion. Our analysis may be helpful to understand microscopic basis of interaction between proteins and ionic species.





Similar content being viewed by others
References
Gale PA, Perez-Tomas R, Quesada R (2013) Anion transporters and biological systems. Acc Chem Res 46(12):2801–2813
Gale PA, Busschaert N, Haynes CJ, Karagiannidis LE, Kirby IL (2014) Anion receptor chemistry: highlights from 2011 and 2012. Chem Soc Rev 43(1):205–241
Caballero A, Zapata F, Beer PD (2013) Interlocked host molecules for anion recognition and sensing. Coord Chem Rev 257(17–18):2434–2455
Gale PA (2011) From anion receptors to transporters. Acc Chem Res 44(3):216–226
Duke RM, Veale EB, Pfeffer FM, Kruger PE, Gunnlaugsson T (2010) Colorimetric and fluorescent anion sensors: an overview of recent developments in the use of 1,8-naphthalimide-based chemosensors. Chem Soc Rev 39(10):3936–3953
Elmes RB, Jolliffe KA (2015) Anion recognition by cyclic peptides. Chem Comm 51(24):4951–4968
Kral V, Rusin O, Shishkanova T, Volf R, Matejka P, Volka K (1999) Anion binding: from supramolecules to sensors. Chem Listy 93(9):546–553
Denessiouk KA, Johnson MS, Denesyuk AI (2005) Novel CαNN structural motif for protein recognition of phosphate ions. J Mol Biol 345(3):611–629
Chakrabarti P (1993) Anion binding sites in protein structures. J Mol Biol 234(2):463–482
Watson JD, Milner-White EJ (2002) A novel main-chain anion-binding site in proteins: the nest. A particular combination of phi, psi values in successive residues gives rise to anion-binding sites that occur commonly and are found often at functionally important regions. J Mol Biol 315(2):171–182
Milner-White EJ, Nissink JW, Allen FH, Duddy WJ (2004) Recurring main-chain anion-binding motifs in short polypeptides: nests. Acta Crystallogr 60:1935–1942
Wintjens R, Rooman M (1996) Structural classification of HTH DNA-binding domains and protein-DNA interaction modes. J Mol Biol 262(2):294–313
Sheet T, Supakar S, Banerjee R (2013) Conformational preference of ‘CαNN’ short peptide motif towards recognition of anions. PLoS ONE 8(3):e57366
Sheet T, Banerjee R (2016) The ‘CαNN’ motif: an intrinsic lover of sulfate and phosphate ions. RSC Adv 6(59):54129–54141
Sheet T, Ghosh S, Pal D, Banerjee R (2017) Computational design of model scaffold for anion recognition based on the ‘CαNN’ motif. Biopolymers 108(1):e22921
Sheet T, Banerjee R (2010) Sulfate ion interaction with ‘anion recognition’ short peptide motif at the N-terminus of an isolated helix: a conformational landscape. J Struct Biol 171(3):345–352
Patra P, Ghosh M, Banerjee R, Chakrabarti J (2017) Anion induced conformational preference of CαNN motif residues in functional proteins. Proteins 85(12):2179–2190
Kagawa H, Mori K (1999) Molecular orbital study of the interaction between MgATP and the myosin motor domain: the highest occupied molecular orbitals indicate the reaction site of ATP hydrolysis. J Phys Chem B 103(34):7346–7352
Blomberg MRA, Siegbahn PEM (2001) A quantum chemical approach to the study of reaction mechanisms of redox-active metalloenzymes. J Phys Chem 105(39):9375–9386
Sikdar S, Ghosh M, De Raychaudhury M, Chakrabarti J (2015) Quantum chemical studies on stability and chemical activities in calcium ion bound calmodulin loops. J Phys Chem B 119(46):14652–14659
Sikdar S, Ghosh M, De Raychaudhury M, Chakrabarti J (2014) Quantum chemical studies on the role of residues in calcium ion binding to calmodulin. Chem Phys Lett 605:103–107
Fukui K (1982) Role of frontier orbitals in chemical reactions. Science 218(4574):747–754
Yao M, Ose T, Sugimoto H, Horiuchi A, Nakagawa A, Wakatsuki S, Yokoi D, Murakami T, Honma M, Tanaka I (2000) Crystal structure of 1-aminocyclopropane-1-carboxylate deaminase from Hansenula saturnus. J Biol Chem 275(44):34557–34565
Dormitzer PR, Sun ZY, Wagner G, Harrison SC (2002) The rhesus rotavirus VP4 sialic acid binding domain has a galectin fold with a novel carbohydrate binding site. EMBO J 21(5):885–897
Symmons MF, Jones GH, Luisi BF (2000) A duplicated fold is the structural basis for polynucleotide phosphorylase catalytic activity, processivity, and regulation. Structure 8(11):1215–1226
Atwell S, Ultsch M, De Vos AM, Wells JA (1997) Structural plasticity in a remodeled protein-protein interface. Science 278(5340):1125–1128
Pieper U, Kapadia G, Mevarech M, Herzberg O (1998) Structural features of halophilicity derived from the crystal structure of dihydrofolate reductase from the Dead Sea halophilic archaeon, Haloferax volcanii. Structure 6(1):75–88
Lepsik M, Field MJ (2007) Binding of calcium and other metal ions to the EF-hand loops of calmodulin studied by quantum chemical calculations and molecular dynamics simulations. J Phys Chem B 111(33):10012–10022
Wu EL, Mei Y, Han K, Zhang JZ (2007) Quantum and molecular dynamics study for binding of macrocyclic inhibitors to human alpha-thrombin. Biophys J 92(12):4244–4253
He X, Zhang JZ (2006) The generalized molecular fractionation with conjugate caps/molecular mechanics method for direct calculation of protein energy. J Chem Phys 124(18):184703
Zhang DW, Chen XH, Zhang JZ (2003) Molecular caps for full quantum mechanical computation of peptide-water interaction energy. J Comput Chem 24(15):1846–1852
Brooks BR, Brooks CL III, Mackerell AD Jr, Nilsson L, Petrella RJ, Roux B, Won Y, Archontis G, Bartels C, Boresch S, Caflisch A, Caves L, Cui Q, Dinner AR, Feig M, Fischer S, Gao J, Hodoscek M, Im W, Kuczera K, Lazaridis T, Ma J, Ovchinnikov V, Paci E, Pastor RW, Post CB, Pu JZ, Schaefer M, Tidor B, Venable RM, Woodcock HL, Wu X, Yang W, York DM, Karplus M (2009) CHARMM: the biomolecular simulation program. J Comput Chem 30(10):1545–1614
Blochl PE (1994) Projector augmented-wave method. Phys Rev B 50(24):17953–17979
Kresse G, Hafner J (1993) Ab-initio molecular-dynamics for open-shell transition-metals. Phys Rev B 48(17):13115–13118
Kresse G, Joubert D (1999) From ultrasoft pseudopotentials to the projector augmented-wave method. Phys Rev B 59(3):1758–1775
Car R, Parrinello M (1985) Unified approach for molecular dynamics and density-functional theory. Phys Rev Lett 55(22):2471–2474
Henkelman G, LaBute MX, Tung CS, Fenimore PW, McMahon BH (2005) Conformational dependence of a protein kinase phosphate transfer reaction. Proc Natl Acad Sci USA 102(43):15347–15351
Adhikari P, Wen AM, French RH, Parsegian VA, Steinmetz NF, Podgornik R, Ching WY (2014) Electronic structure, dielectric response, and surface charge distribution of RGD (1FUV) peptide. Sci Rep 4:5605
Silvestrelli PL, Parrinello M (1999) Structural, electronic, and bonding properties of liquid water from first principles. J Chem Phys 111(8):3572–3580
Kresse G, Hafner J (1993) Abinitio molecular-dynamics for liquid-metals. Phys Rev B 47(1):558–561
Kresse G, Furthmuller J (1996) Efficient iterative schemes for ab initio total-energy calculations using a plane-wave basis set. Phys Rev B 54(16):11169–11186
The PyMOL Molecular Graphics System. Version 2.0 edn. Schrodinger, LLC
Gaussian 03, Revision C, Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Montgomery JA Jr, Vreven T, Kudin KN, Burant JC, Millam JM, Iyengar SS, Tomasi J, Barone V, Mennucci B, Cossi M, Scalmani G, Rega N, Petersson GA, Nakatsuji H, Hada M, Ehara M, Toyota K, Fukuda R, Hasegawa J, Ishida M, Nakajima T, Honda Y, Kitao O, Nakai H, Klene M, Li X, Knox JE, Hratchian HP, Cross JB, Bakken V, Adamo C, Jaramillo J, Gomperts R, Stratmann RE, Yazyev O, Austin AJ, Cammi R, Pomelli C, Ochterski JW, Ayala PY, Morokuma K, Voth GA, Salvador P, Dannenberg JJ, Zakrzewski VG, Dapprich S, Daniels AD, Strain MC, Farkas O, Malick DK, Rabuck AD, Raghavachari K, Foresman JB, Ortiz JV, Cui Q, Baboul AG, Clifford S, Cioslowski J, Stefanov BB, Liu G, Liashenko A, Piskorz P, Komaromi I, Martin RL, Fox DJ, Keith T, Al-Laham MA, Peng CY, Nanayakkara A, Challacombe M, Gill PMW, Johnson B, Chen W, Wong MW, Gonzalez C, Pople JA (2004) Gaussian, Inc., Wallingford CT
Trado-Rives J, Jorgensen WL (2008) Performance of B3LYP density functional methods for a large set of organic molecules. J Chem Theory Comput 4(2):297–306
Siegbahn PE, Borowski T (2006) Modeling enzymatic reactions involving transition metals. Acc Chem Res 39:729–738
Blomberg MR, Siegbahn PE (2006) Quantum chemistry applied to the mechanisms of transition metal containing enzymes-cytochrome c oxidase, a particularly challenging case. J Comput Chem 27:1373–1384
Acknowledgements
PP thanks to Dr. Manas Mondal for helping in VASP calculation. RB and PP thank the TEQIP-III, MAKAUT, WB and DBT-BIF for funding. PP is also thankful to SNBNCBS, Kolkata for giving the computational facility. JC thanks DST for financial support.
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Patra, P., Ghosh, M., Banerjee, R. et al. Quantum chemical studies on anion specificity of CαNN motif in functional proteins. J Comput Aided Mol Des 32, 929–936 (2018). https://doi.org/10.1007/s10822-018-0157-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10822-018-0157-3