Abstract
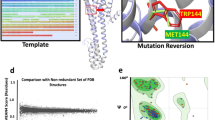
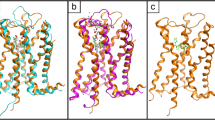
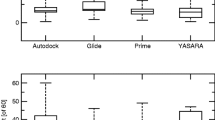
The allosteric modulation of G protein-coupled receptors (GPCRs) by sodium ions has received considerable attention as crystal structures of several receptors, in their inactive conformation, show a Na+ ion bound to specific residues which, in the human A2A adenosine receptor (hA2A AR), are Ser913.39, Trp2466.48, Asn2807.45, and Asn2847.49. A cluster of water molecules completes the coordination of the sodium ion in the putative allosteric site. It is absolutely consolidated that the progress made in the field of GPCRs structural determination has increased the adoption of docking-driven approaches for the identification or the optimization of novel potent and selective ligands. Despite the extensive use of docking protocols in virtual screening approaches, to date, almost any of these studies have been carried out without taking into account the presence of the sodium cation and its first solvation shell in the putative allosteric binding site. In this study, we have focused our attention on determining how the presence of sodium ion binding and additionally its first hydration sphere, in hA2AAR could influence the ligand positioning accuracy during molecular docking simulations for most of the available resting and activated hA2A AR crystal structures, using DockBench as a comparative benchmarking tool and implementing a new correlation coefficient (EM). This work provides indications on the evidence that the posing performance (accuracy and/or precision) of the docking protocols in reproducing the crystallographic poses of different hA2A AR antagonists is generally increased in the presence of the sodium cation and its first solvation shell, in agreement with experimental observations. Consequently, the inclusion of sodium ion and its first solvation shell should be considered in order to facilitate the selection of new potential ligands in all molecular docking-based virtual screening protocols that aim to find novel GPCRs antagonists and inverse agonists.





Similar content being viewed by others
References
Jacobson KA (2009) Introduction to adenosine receptors as therapeutic targets. Handb Exp Pharmacol 193:1–24
Liu W, Chun E, Thompson AA et al (2012) Structural basis for allosteric regulation of GPCRs by sodium ions. Science 337:232–236
Ballesteros JA, Weinstein H (1995) [19] Integrated methods for the construction of three-dimensional models and computational probing of structure-function relations in G protein-coupled receptors. Recept Mol Biol. https://doi.org/10.1016/S1043-9471(05)80049-7
Massink A, Gutiérrez-de-Terán H, Lenselink EB et al (2015) Sodium ion binding pocket mutations and adenosine A2A receptor function. Mol Pharmacol 87:305–313
Segala E, Guo D, Cheng RKY et al (2016) Controlling the dissociation of ligands from the adenosine A2A receptor through modulation of salt bridge strength. J Med Chem 59:6470–6479
Cheng RKY, Segala E, Robertson N et al (2017) Structures of human A1and A2AAdenosine receptors with xanthines reveal determinants of selectivity. Structure 25:1275–1285.e4
Rucktooa P, Cheng RKY, Segala E et al (2018) Towards high throughput GPCR crystallography: in meso soaking of adenosine A2AReceptor CRYSTALS. Sci Rep 8:41
Batyuk A, Galli L, Ishchenko A et al (2016) Native phasing of X-ray free-electron laser data for a G protein-coupled receptor. Sci Adv 2:e1600292
Weinert T, Olieric N, Cheng R et al (2017) Serial millisecond crystallography for routine room-temperature structure determination at synchrotrons. Nat Commun 8:542
Broecker J, Morizumi T, Ou W-L et al (2018) High-throughput in situ X-ray screening of and data collection from protein crystals at room temperature and under cryogenic conditions. Nat Protoc 13:260–292
Gutiérrez-de-Terán H, Massink A, Rodríguez D et al (2013) The role of a sodium ion binding site in the allosteric modulation of the A(2A) adenosine G protein-coupled receptor. Structure 21:2175–2185
Selvam B, Shamsi Z, Shukla D (2018) Universality of the sodium ion binding mechanism in class A G-protein-coupled receptors. Angew Chem Int Ed 57:3048–3053
Katritch V, Fenalti G, Abola EE et al (2014) Allosteric sodium in class A GPCR signaling. Trends Biochem Sci 39:233–244
Massink A, Louvel J, Adlere I et al (2016) 5′-substituted amiloride derivatives as allosteric modulators binding in the sodium ion pocket of the adenosine A2A receptor. J Med Chem 59:4769–4777
Ye L, Neale C, Sljoka A et al (2018) Mechanistic insights into allosteric regulation of the A2A adenosine G protein-coupled receptor by physiological cations. Nat Commun 9:1372
Ciancetta A, Sabbadin D, Federico S et al (2015) Advances in computational techniques to study GPCR-ligand recognition. Trends Pharmacol Sci 36:878–890
Cuzzolin A, Sturlese M, Malvacio I et al (2015) DockBench: an integrated informatic platform bridging the gap between the robust validation of docking protocols and virtual screening simulations. Molecules 20:9977–9993
Salmaso V, Sturlese M, Cuzzolin A, Moro S (2016) DockBench as docking selector tool: the lesson learned from D3R Grand Challenge 2015. J Comput Aided Mol Des 30:773–789
Hollenstein K, de Graaf C, Bortolato A et al (2014) Insights into the structure of class B GPCRs. Trends Pharmacol Sci 35:12–22
Mason JS, Bortolato A, Congreve M, Marshall FH (2012) New insights from structural biology into the druggability of G protein-coupled receptors. Trends Pharmacol Sci 33:249–260
Tautermann CS (2014) GPCR structures in drug design, emerging opportunities with new structures. Bioorg Med Chem Lett 24:4073–4079
Congreve M, Marshall F (2010) The impact of GPCR structures on pharmacology and structure-based drug design. Br J Pharmacol 159:986–996
Chemical Computing Group - Citing MOE. https://www.chemcomp.com/Research-Citing_MOE.htm. Accessed 3 Oct 2016
gnuplot homepage. http://gnuplot.sourceforge.net/. Accessed 28 Mar 2018
MarvinSketch. https://docs.chemaxon.com/display/docs/MarvinSketch+Home. Accessed 28 Mar 2018
Pettersen EF, Goddard TD, Huang CC et al (2004) UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612
Berman HM, Westbrook J, Feng Z et al (2000) The protein data bank. Nucleic Acids Res 28:235–242
Labute P (2009) Protonate3D: assignment of ionization states and hydrogen coordinates to macromolecular structures. Proteins 75:187–205
Labute P (2008) The generalized Born/volume integral implicit solvent model: estimation of the free energy of hydration using London dispersion instead of atomic surface area. J Comput Chem 29:1693–1698
Wang J, Wolf RM, Caldwell JW et al (2004) Development and testing of a general amber force field. J Comput Chem 25:1157–1174
Stewart JJP (1989) Optimization of parameters for semiempirical methods I. Method. J Comput Chem 10:209–220
Morris GM, Huey R, Lindstrom W et al (2009) AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem 30:2785–2791
Friesner RA, Banks JL, Murphy RB et al (2004) Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J Med Chem 47:1739–1749
GOLD - The Cambridge Crystallographic Data Centre (CCDC). https://www.ccdc.cam.ac.uk/solutions/csd-discovery/components/gold/. Accessed 12 Mar 2017
Korb O, Stützle T, Exner TE (2009) Empirical scoring functions for advanced protein-ligand docking with PLANTS. J Chem Inf Model 49:84–96
Ruiz-Carmona S, Alvarez-Garcia D, Foloppe N et al (2014) rDock: a fast, versatile and open source program for docking ligands to proteins and nucleic acids. PLoS Comput Biol 10:e1003571
O’Boyle NM, Banck M, James CA et al (2011) Open Babel: an open chemical toolbox. J Cheminform 3:33
Gasteiger J, Marsili M (1980) Iterative partial equalization of orbital electronegativity—a rapid access to atomic charges. Tetrahedron 36:3219–3228
Trott O, Olson AJ (2010) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 31:455–461
Li P, Song LF, Merz KM (2015) Systematic parameterization of monovalent ions employing the nonbonded model. J Chem Theory Comput 11:1645–1657
Ciancetta A, Cuzzolin A, Moro S (2014) Alternative quality assessment strategy to compare performances of GPCR-ligand docking protocols: the human adenosine A(2A) receptor as a case study. J Chem Inf Model 54:2243–2254
Hino T, Arakawa T, Iwanari H et al (2012) G-protein-coupled receptor inactivation by an allosteric inverse-agonist antibody. Nature 482:237–240
Complex with ZM241385 and the xanthines XAC and caffeine. Structure 19:1283–1293
Jaakola V-P, Griffith MT, Hanson MA et al (2008) The 2.6 angstrom crystal structure of a human A2A adenosine receptor bound to an antagonist. Science 322:1211–1217
Glukhova A, Thal DM, Nguyen AT et al (2017) Structure of the adenosine A1 receptor reveals the basis for subtype selectivity. Cell 168:867–877.e13
Congreve M, Andrews SP, Doré AS et al (2012) Discovery of 1,2,4-triazine derivatives as adenosine A(2A) antagonists using structure based drug design. J Med Chem 55:1898–1903
Melnikov I, Polovinkin V, Kovalev K et al (2017) Fast iodide-SAD phasing for high-throughput membrane protein structure determination. Sci Adv 3:e1602952
Selvam B, Shamsi Z, Shukla D (2018) Universality of the sodium ion binding mechanism in class A G-protein-coupled receptors. Angew Chem Int Ed Engl 57:3048–3053
Mooij WTM, Verdonk ML (2005) General and targeted statistical potentials for protein–ligand interactions. Proteins 61:272–287
Cheng T, Li X, Li Y et al (2009) Comparative assessment of scoring functions on a diverse test set. J Chem Inf Model 49:1079–1093
Li X, Li Y, Cheng T et al (2010) Evaluation of the performance of four molecular docking programs on a diverse set of protein–ligand complexes. J Comput Chem 31:2109–2125
Li Y, Han L, Liu Z, Wang R (2014) Comparative assessment of scoring functions on an updated benchmark: 2. Evaluation methods and general results. J Chem Inf Model 54:1717–1736
Pagadala NS, Syed K, Tuszynski J (2017) Software for molecular docking: a review. Biophys Rev 9:91–102
Lenselink EB, Beuming T, Sherman W et al (2014) Selecting an optimal number of binding site waters to improve virtual screening enrichments against the adenosine A2A receptor. J Chem Inf Mod 54(6):1737–1746
Acknowledgements
MMS lab is very grateful to Chemical Computing Group and OpenEye for the scientific and technical partnership. MMS lab gratefully acknowledges the support of NVIDIA Corporation with the donation of the Titan Xp GPU used for this research.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Margiotta, E., Deganutti, G. & Moro, S. Could the presence of sodium ion influence the accuracy and precision of the ligand-posing in the human A2A adenosine receptor orthosteric binding site using a molecular docking approach? Insights from Dockbench. J Comput Aided Mol Des 32, 1337–1346 (2018). https://doi.org/10.1007/s10822-018-0174-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10822-018-0174-2