Abstract
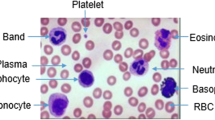
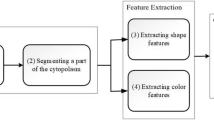
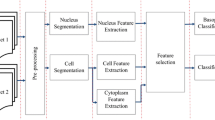
Peripheral Blood Smear analysis plays a vital role in diagnosis of many diseases such as leukemia, anemia, malaria, lymphoma and infections. Unusual variations in color, shape and size of blood cells indicate abnormal condition. We used a total of 117 images from Leishman stained peripheral blood smears acquired at a magnification of 100X. In this paper we present a robust image processing algorithm for detection of nuclei and classification of white blood cells based on features of the nuclei. We used novel image enhancement method to manage illumination variations and TissueQuant method to manage color variations for the detection of nuclei. Dice similarity coefficient of 0.95 was obtained for nucleus detection. We also compared the proposed method with a state-of-the-art method and the proposed method was found to be better. Shape and texture features of the detected nuclei were used for classifying white blood cells. We considered classification of WBCs using two approaches such as 5-class and cell-by-cell approaches using neural network and hybrid-classifier respectively. We compared the results of both the approaches for classification of white blood cells. Cell-by-cell approach offered 1.4% higher sensitivity in comparison with the 5-class approach. We obtained an accuracy of 100% for lymphocyte and basophil detection. Hence, we conclude that lymphocytes and basophils can be accurately detected even when the analysis is limited to the features of nuclei whereas, accurate detection of other types of WBCs will require analysis of the cytoplasm too.








Similar content being viewed by others
References
Adnan, K., IBCIS: intelligent blood cell identification system. Prog. Nat. Sci. 18:1309–1314, 2008.
Maity, M., Mungle, T., Dhane, D., Maiti, A. K., and Chakraborty, C., An ensemble rule learning approach for automated morphological classification of erythrocytes. J. Med. Syst. 41(4):56, 2017. https://doi.org/10.1007/s10916-017-0691-x.
Prasad, K., and Prabhu, G. K., Image analysis tools for evaluation of microscopic views of immunohistochemically stained specimen in medical research–a review. J. Med. Syst. 36(4):2621–2631, 2012. https://doi.org/10.1007/s10916-011-9737-7.
Ahmad, A., Asif, A., Rajpoot, N., Arif, M., and Minhas, F. U. A. A., Correlation filters for detection of cellular nuclei in histopathology images. J. Med. Syst. 42(1):7, 2017. https://doi.org/10.1007/s10916-017-0863-8.
Madhloom, H. T., Kareem, S. A., Ariffin, H., Zaidan, A. A., Alanazi, H. O., and Zaidan, B. B., An automated white blood cell nucleus localization and segmentation using image arithmetic and automatic threshold. J. Appl. Sci. 10(11):959–966, 2010.
Neelam, S., Ramakrishnan, A. G., Automation of differential blood count. Proc. IEEE Conference on Convergent Technologies for the Asia-Pacific Region 2:547–551, 2003. https://doi.org/10.1109/TENCON.2003.1273221
Nisha, R., Bryan, D., Mohammed, E. S., and Tolga, T., Isolation and two-step classification of normal white blood cells in peripheral blood smears. J. Pathol. Informatics 3(1):10, 2012. https://doi.org/10.4103/2153-3539.93895.
Salim, A., Emel, O., and Cigdem Gunduz, D., A color and shape based algorithm for segmentation of white blood cells in peripheral blood and bone marrow images. Cytometry A 85:480–490, 2014. https://doi.org/10.1002/cyto.a.22457.
Hayan, T. M., Sameem Abdul, K., and Hany, A., An image processing application for the localization and segmentation of lymphoblast cell using peripheral blood images. J. Med. Syst. 36(4):2149–2158, 2012. https://doi.org/10.1007/s10916-011-9679-0.
Hiremath, P. S., Parashuram, B., and Sai, G., Automated identification and classification of white blood cells (leukocytes) in digital microscopic images. IJCA Special Issue RTIPPR 2:59–63, 2010.
Jaroonrut, P., and Charnchai, P., Segmentation of white blood cells and comparison of cell morphology by linear and naive Bayes classifiers. Biomed. Eng. Online 14(63):63, 2015. https://doi.org/10.1186/s12938-015-0037-1.
Sawsan, F. B., Ahmed, M. D., Hany, A. T., and Samir, I. S., Segmentation and classification of white blood cells. Proc. IEEE Int. Conf. Acoust. Speech Signal Process 4:2259–2261, 2000. https://doi.org/10.1109/ICASSP.2000.859289.
Sedat, N., Deniz, K., Tuncay, E., Murat Husnu, S., Osman, K., and Yavuz, E., Automatic segmentation, counting, size determination and classification of white blood cells. J. Meas. 55:58–65, 2014. https://doi.org/10.1016/j.measurement.2014.04.008.
Bakht, A., Rashid Jalal, Q., Zahoor, J., and Taj Ali, K., Color based segmentation of white blood cells in blood photomicrographs using image quantization. Res. J. Recent Sci. 3(4):34–39, 2014.
Seyed Hamid, R., and Hamid Soltanian, Z., Automatic recognition of five types of white blood cells in peripheral blood. Comput. Med. Imaging Graph. 35(4):333–343, 2011. https://doi.org/10.1016/j.compmedimag.2011.01.003.
Mathur, A., Tripathi, A. S., and Kuse, M., Scalable system for classification of white blood cells from Leishman stained blood stain images. J. Pathol. Informatics 4(2):15, 2013. https://doi.org/10.4103/2153-3539.109883.
Der-Chen, H., Kun Ding, H., and Yung Kuan, C., A computer assisted method for leukocyte nucleus segmentation and recognition in blood smear images. J. Syst. Softw. 85:2104–2118, 2012. https://doi.org/10.1016/j.jss.2012.04.012.
Dipti Prasad, M., Nilanjan, R., and Scott, T. A., Level set analysis for leukocyte detection and tracking. IEEE Trans. Image Process. 13(4):562–572, 2004. https://doi.org/10.1109/TIP.2003.819858.
Keerthana, P., Bhagath, K., Marx, C., and Gopalakrishna, P., Applications of TissueQuant a color intensity quantification tool for medical research. Comput. Methods Prog. Biomed. 106(1):27–36, 2012. https://doi.org/10.1016/j.cmpb.2011.08.004.
Matthew, S., Finding dents in a blobby shape, 2013. URL https://syntacticsalt.com/2013/01/11/finding-dents-in-an-blobby-shape/
Steven, G. J., Notes on FFT-based differentiation, MIT applied mathematics pp. 1–11, 2011. URL http://math.mit.edu/stevenj/fft-deriv.pdf
Haralick, R. M., Shanmugam, K., and Dinstein, I., Textural features for image classification. IEEE Trans. Syst. Man Cybern. SMC-3(6):610–621, 1973. https://doi.org/10.1109/TSMC.1973.4309314.
Keerthana, P., Jan, W., Udayakrishna, M. B., Raviraja, V. A., and Gopalakrishna, K. P., Image analysis approach for development of a decision support system for detection of malaria parasites in thin blood smear images. J. Digit. Imaging 25(4):542–549, 2012. https://doi.org/10.1007/s10278-011-9442-6.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest. This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Additional information
This article is part of the Topical Collection on Image & Signal Processing
Rights and permissions
About this article
Cite this article
Hegde, R.B., Prasad, K., Hebbar, H. et al. Development of a Robust Algorithm for Detection of Nuclei and Classification of White Blood Cells in Peripheral Blood Smear Images. J Med Syst 42, 110 (2018). https://doi.org/10.1007/s10916-018-0962-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10916-018-0962-1