Abstract
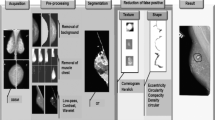
The present study introduces an efficient algorithm for automatic segmentation and detection of mass present in the mammograms. The problem of over and under-segmentation of low-contrast mammographic images has been solved by applying preprocessing on original mammograms. Subtraction operation performed between enhanced and enhanced inverted mammogram significantly highlights the suspicious mass region in mammograms. The segmentation accuracy of suspicious region has been improved by combining wavelet transform and fast fuzzy c-means clustering algorithm. The accuracy of mass segmentation has been quantified by means of Jaccard coefficients. Better sensitivity, specificity, accuracy, and area under the curve (AUC) are observed with support vector machine using radial basis kernel function. The proposed algorithm is validated on Mini-Mammographic Image Analysis Society (MIAS) and Digital Database for Screening Mammography (DDSM) datasets. Highest 91.76% sensitivity, 96.26% specificity, 95.46% accuracy, and 96.29% AUC on DDSM dataset and 94.63% sensitivity, 92.74% specificity, 92.02% accuracy, and 95.33% AUC on MIAS dataset are observed. Also, shape analysis of mass is performed by using moment invariant and Radon transform based features. The best results are obtained with Radon based features and achieved accuracies for round, oval, lobulated, and irregular shape of mass are 100%, 70%, 64%, and 96%, respectively.










Similar content being viewed by others
References
American College Radiology (1998) BI-RADS Committee. Breast imaging reporting and data system. American College of Radiology, Reston
Bellotti R, De Carlo F, Tangaro S, Gargano G, Maggipinto G, Castellano M, Massafra R, Cascio D, Fauci F, Magro R, Raso G (2006) A completely automated CAD system for mass detection in a large mammographic database. Med phy 33:3066–3075. doi:10.1118/1.2214177
Bocchi L, Nori J (2007) Shape analysis of micro-calcifications using radon transform. Med Eng Phy 29:691–698. doi:10.1016/j.medengphy.2006.07.012
Cao A, Song Q, Yang X, Wang L (2004) Breast mass segmentation based on information theory. In: Pattern Recognition, 2004. ICPR 2004. Proceedings of the 17th IEEE International Conference vol. 3, pp 758–761. Doi:10.1016/j.cviu.2007.07.004
Cascio D, Fauci F, Magro R, Raso G, Bellotti R, De Carlo F, Tangaro S, De Nunzio G, Quarta M, Forni G et al (2006) Mammogram segmentation by contour searching and mass lesions classification with neural network. IEEE Trans Nucl Sci 53:2827–2833. doi:10.1109/TNS.2006.878003
Che FN, Fairhust MC, Wells CP, Hanson M (1996) Evaluation of a two-stage model for detection of abnormalities in digital mammograms. In: IEEE Colloquium Digest Mammography, pp 13–1. Doi: 10.1049/ic:19960496
Cheetham AH, Hazel JE (1969) Binary (presence-absence) similarity coefficients. J Paleontol 43(5):1130–1136
Christoyianni I, Dermatas E, Kokkinakis G (2000) Fast detection of masses in computer-aided mammography. Sig Proc Mag 17:54–64. doi:10.1109/79.814646
Constantinidis AS, Fairhust MC, Deravi F, Hanson M, Wells CP, Chapman-Jones C (1999) Evaluating classification strategies for detection of circumscribed masses in digital mammograms. In: International Conference on Image Processing and its Application. pp 435–439. Doi:10.1049/cp:19990359
Cortes C, Vapnik V (1995) Support-vector networks. Mach Learn 20(3):273–297
Deans SR (2007) The Radon transform and some of its applications. Courier Corporation, North Chelmsford
Do Nascimento MZ, Martins AS, Neves LA, Ramos RP, Flores EL, Carrijo GA (2013) Classification of masses in mammographic image using wavelet domain features and polynomial classifier. Expert Sys Appl 40:6213–6221. doi:10.1016/j.eswa.2013.04.036
Dominguez AR, Nandi AK (2009) Toward breast cancer diagnosis based on automated segmentation of masses in mammograms. Pattern Recog 42:1138–1148. doi:10.1016/j.patcog.2008.08.006
Ganesan K, Acharya UR, Chua CK, Min LC, Abraham KT, Ng KH (2013) Computer-aided breast cancer detection using mammograms: a review. IEEE Rev Biomed Eng 6:77–98. doi:10.1109/ICoCS.2014.7060995
Gonzalez RC, Woods RE (2008) Digital Image Processing, 3rd edn. Pearson Prentice Hall, Upper Saddle River
Haralick RM, Shanmugam K, Dinstein IH (1973) Textural features for image classification. IEEE Trans Syst Man Cybern 6:610–621. doi:10.1109/TSMC.1973.4309314
Hu MK (1962) Visual pattern recognition by moment invariants. IRE Trans Inf Theory 8:179–187
Hu K, Gao X, Li F (2011) Detection of suspicious lesions by adaptive thresholding based on multi-resolution analysis in mammograms. IEEE Trans Instrum Meas 60:462–472. doi:10.1109/TIM.2010.2051060
Huo Z, Giger ML, Vyborny CJ, Bick U, Lu P, Wolverton DE, Schmidt RA (1995) Analysis of spiculation in the computerized classification of mammographic masses. Med Phys 22:1569–1579. doi:10.1118/1.597626
Jen CC, Yu SS (2015) Automatic detection of abnormal mammograms in mammographic images. Expert Syst Appl 42:3048–3055. doi:10.1016/j.eswa.2014.11.061
Jolliffe I (2002) Principal component analysis. John Wiley & Sons, Ltd., New York
Kom G, Tiedeu A, Kom M (2007) Automated detection of masses in mammograms by local adaptive thresholding. Comput Biol Med 37:37–48. doi:10.1016/j.compbiomed.2005.12.004
Kupinski MA, Giger ML (1998) Automated seeded lesion segmentation on digital mammograms. IEEE Trans Med Imag 17:510–517. doi:10.1109/42.730396
Kurt B, Nabiyev VV, Turhan K (2014) A novel automatic suspicious mass regions identification using Havrda & Charvat entropy and Otsu's N thresholding. Comput Methods Prog Biomed 114:349–360. doi:10.1016/j.cmpb.2014.02.014
Llado X, Oliver A, Freixenet J, Mart R, Mart J (2009) A textural approach for mass false positive reduction in mammography. Comput Med Imaging Graph 33:415–422. doi:10.1016/j.compmedimag.2009.03.007
Materka A, Strzelecki M et al (1998) Texture analysis methods–a review. Technical University of Lodz, Institute of Electronics, COST B11 Report, Brussels, pp 9–11
Muramatsu C, Hara T, Endo T, Fujita H (2016) Breast mass classification on mammograms using radial local ternary patterns. Comput Biol Med 72:43–53. doi:10.1016/j.compbiomed.2016.03.007
Ng SL, Bischof WF (1992) Automated detection and classification of breast tumors. Comput Biomed Res 25:218–237. doi:10.1016/0010-4809(92)90040-H
Nguyen TM, Rangayyan RM (2006) Shape analysis of breast masses in mammograms via the fractal dimension. In: Engineering in Medicine and Biology Society, 2005. IEEE-EMBS 2005. 27th Annual International Conference pp 3210–3213. doi:10.1109/IEMBS.2005.1617159
Nunes AP, Silva AC, de Paiva AC (2010) Detection of masses in mammographic images using geometry, Simpson’s diversity index and SVM. Int J Signal Imaging Syst Eng 3:43–51. doi:10.1504/IJSISE.2010.034631
Ojala T, Pietikainen M, Maenpaa T (2002) Multi-resolution gray-scale and rotation invariant texture classification with local binary patterns. IEEE Trans Pattern Anal Mach Intell 24:971–987. doi:10.1109/TPAMI.2002.1017623
Oliveira Martins L, Silva AC, De Paiva AC, Gattass M (2009) Detection of breast masses in mammogram images using growing neural gas algorithm and Ripley’s K function. J Signal Process Syst 55:77–90. doi:10.1007/s11265-008-0209-3
Oliver A, Freixenet J, Martí J, Pérez E, Pont J, Denton ERE, Zwiggelaar R (2010) A review of automatic mass detection and segmentation in mammographic images. Med Image Anal 14:87–110. doi:10.1016/j.media.2009.12.005
Pal NR, Bezdek JC (1995) On cluster validity for the fuzzy c-means model. IEEE Trans Fuzzy Syst 3:370–379. doi:10.1109/91.413225
Pereira DC, Ramos RP, Do Nascimento MZ (2014) Segmentation and detection of breast cancer in mammograms combining wavelet analysis and genetic algorithm. Comput Methods Prog Biomed 114:88–10. doi:10.1016/j.cmpb.2014.01.014
Petrick N, Chan HP, Sahiner B, Wei D (1996a) An adaptive density-weighted contrast enhancement filter for mammographic breast mass detection. IEEE Trans Med Imaging 15:59–67. doi:10.1109/42.481441
Petrick N, Chan HP, Wei D, Sahiner B, Helvie MA, Adler DD (1996b) Automated detection of breast masses on mammograms using adaptive contrast enhancement and texture classification. Med Phys 23:1685–1696. doi:10.1118/1.597756
Rose C, Turi D, Williams A, Wolstencroft K, Taylor C (2006) Web services for the ddsm and digital mammography research. In: Digital Mammography pp 376–383
Sampaio WB et al (2011) Detection of masses in mammogram images using CNN, geostatistic functions and SVM. Comput Biol Med 41:653–664. doi:10.1016/j.compbiomed.2005.12.004
de Sampaio WB, Silva AC, de Paiva AC, Gattass M (2015) Detection of masses in mammograms with adaption to breast density using genetic algorithm, phylogenetic trees, LBP and SVM. Expert Syst Appl 42:8911–8928. doi:10.1016/j.eswa.2015.07.046
Sharma S, Khanna P (2015) Computer-aided diagnosis of malignant mammograms using Zernike moments and SVM. J Digit Imaging 28:77–90. doi:10.1007/s10278-014-9719-7
Suckling J, Parker J, Dance D, Astley S, Hutt I, Boggis C, Ricketts I, Stamatakis E, Cerneaz N, Kok S, Taylor P (1994) The mammographic image analysis society digital mammogram database. Excerpta Med Int Congr Ser 1069:375–378
Surendiran B, Vadivel A (2012) Mammogram mass classification using various geometric shape and margin features for early detection of breast cancer. Int J Med Eng Inf 4:36–54. doi:10.1504/IJMEI.2012.045302
Tai SC, Chen ZS, Tsai WT (2014) An automatic mass detection system in mammograms based on complex texture features. IEEE J Biomed Health Inform 18:618–627. doi:10.1109/JBHI.2013.2279097
Tang X (1998) Texture information in run length matrices. IEEE Trans Image Process 7:1602–1609. doi:10.1109/83.725367
Wang D, Shi L, Heng PA (2009) Automatic detection of breast cancers in mammograms using structured support vector machines. Neurocomputing 72:3296–3302. doi:10.1016/j.neucom.2009.02.015
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kashyap, K.L., Bajpai, M.K. & Khanna, P. An efficient algorithm for mass detection and shape analysis of different masses present in digital mammograms. Multimed Tools Appl 77, 9249–9269 (2018). https://doi.org/10.1007/s11042-017-4751-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-017-4751-5