Abstract
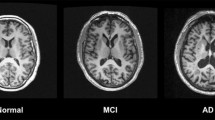
Traditional researches assume that brain functional networks are static during the entire scanning process of functional magnetic resonance image (fMRI) in the resting state. However, it is not difficult to ignore the dynamic interaction patterns of brain regions that essentially change across time. In this study, we take the internal weight information of the brain functional network as a calculation condition of module dividing for brain functional networks. The concept of betweenness efficiency is firstly proposed to improve Girvan-Newman (GN) algorithm for a better module dividing result, and the maximum modularity is used as a criterion to classify the brain functional network modules of normal subjects. The effect of the improved method is verified by controlling subjects, parameters, environment and other conditions. Then, the improved method was utilized to separate the modules in brain functional networks of normal subjects, and the template was used to divide the functional network of Alzheimer’s disease (AD) patients and mild cognitive impairment (MCI) sufferers. The shortest path length of each module is calculated, and the experimental results are compared with the original GN and weighted GN algorithm improved by Newman. The experimental results demonstrate that, the maximum modularity of the improved method is higher while the dividing effect is better under the same conditions. Meanwhile, the conclusion is consistent with the existing research results when the proposed method is applicable to the analysis of the shortest path length. These results illustrate the viewpoint that the proposed method of module dividing is feasible in the analysis of modular structure of brain functional network.








Similar content being viewed by others
References
Atangana A, Liu AJ, Lu ZY (2018) Application of stationary wavelet entropy in pathological brain detection. Multimed Tools Appl 77(3):3701–3714
Chang CT, Glover GH (2010) Time-frequency dynamics of resting-state brain connectivity measured with fMRI. Neurolmage 50(1):81–98
Chen XB, Zhang H, Zhang LC, Shen C, Lee SW, Shen DG (2017) Extraction of dynamic functional connectivity from brain grey matter and white matter for MCI classification. Hum Brain Mapp 38(10):5019–5034
Chen XB, Zhang H, Lee SW, Shen DG (2017) Hierarchical high-order functional connectivity networks and selective feature fusion for MCI classification. Neuroinformatics 15(3):271–284
Echávarri C, Aalten P, Uylings H, Jacobs H, Visser P, Gronenschild E, Verhey F, Burgmans S (2011) Atrophy in the parahippocampal gyrus as an early biomarker of Alzheimer’s disease. Brain Struct Funct 215(3–4):265–271
Girvan M, Newman MEJ (2001) Community structure in social and biological networks. Proc Natl Acad Sci 99:7821–7826
Hutchison RM, Womelsdorf T, Gati JS (2013) Resting-state networks show dynamic functional connectivity in awake humans and anesthetized macaques. Hum Brain Mapp 34(9):2154–2177
Jiang YY, Zhu WG, Lu SY, Zhao GH (2018) Exploring a smart pathological brain detection method on Pseudo Zernike moment. Multimed Tools Appl 77(17):22589–22604
Jiao ZQ, Zou L, Cao Y, Qian N, Ma ZH (2014) Effective connectivity analysis of fMRI data based on network motifs. J Supercomput 67(3):809–819
Jiao ZQ, Wang H, Ma K (2016) The connectivity measurement in complex directed networks by motif structure. Int J Sensor Netw 21(3):197–204
Jiao ZQ, Wang H, Ma K, Zou L, Xiang JB (2017) Directed connectivity of brain default networks in resting state using GCA and motif. Front Biosci Landmark 22(10):1634–1643
Jiao ZQ, Wang H, Ma K, Zou L, Xiang JB, Wang SH (2017) Effective connectivity in the default network using granger causal analysis. J Med Imaging Health Inf 7(2):407–415
Jiao ZQ, Ma K, Wang H, Zou L, Xiang JB (2017) Functional connectivity analysis of brain default mode networks using Hamiltonian path. CNS Neurol Disord Drug Targets 16(1):44–50
Jiao ZQ, Ma K, Rong YL, Wang H, Zou L (2017) Adaptive synchronization in small-world networks with Lorenz chaotic oscillators. Int J Sensor Netw 24(2):90–97
Jiao ZQ, Xia ZW, Cai M, Zou L, Xiang JB, Wang SH (2018) Hub recognition for brain functional networks by using multiple-feature combination. Comput Electr Eng 69:740–745
Jiao ZQ, Ma K, Wang H, Zou L, Zhang YD (2018) Research on node properties of resting-state brain functional networks by using node activity and ALFF. Multimed Tools Appl 77(17):22689–22704
Kaiser M (2011) A tutorial in connectome analysis: topological and spatial features of brain networks. NeuroImage 57(3):892–907
Koch W, Teipel S, Mueller S, Benninghoff J, Wagner M, Bokde AL, Hampel H, Coates U, Reiser M, Meindl T (2012) Diagnostic power of default mode network resting state fMRI in the detection of Alzheimer's disease. Neurobiol Aging 33(3):466–478
Laurienti P, Hugenschmidt C, Hayasaka S (2009) Modularity maps reveal community structure in the resting human brain. Nature Publishing Group
Li HJ, Daniels JJ (2015) Social significance of community structure: statistical view. Phys Rev E 91(1):012801
Li HJ, Li HY (2016) Scalably revealing the dynamics of soft community structure in complex networks. J Syst Sci Complex 29(4):1071–1088
Li XJ, Zhang P, Di ZR, Fan W (2008) Community structure in complex networks. J Complex Syst Complexity 5(3):19–42
Li HJ, Wang H, Chen L (2015) Measuring robustness of community structure in complex networks. Europhys Lett 108(6):68009
Li HJ, Bu Z, Li AH, Liu ZD, Shi Y (2016) Fast and accurate mining the community structure: integrating center locating and membership optimization. IEEE Trans Knowl Data Eng 28(9):2349–2362
Li HJ, Bu Z, Li YL, Zhang ZY, Chu YC, Li GJ, Cao J (2018) Evolving the attribute flow for dynamical clustering in signed networks. Chaos, Solitons Fractals 110:20–27
Lord LD, Stevner AB, Deco G (2017) Understanding principles of integration and segregation using whole-brain computational connectomics: implications for neuropsychiatric disorders. Philos Trans 375:2096 20160283
Newman MEJ (2004) Fast algorithm for detecting community structure in networks. Phys Rev E 69(6):066133
Newman MEJ, Girvan M (2004) Finding and evaluating community structure in networks. Phys Rev E 69(2):026113
Nigam S, Shimono M, Ito S, Yeh FC, Timme N, Myroshnychenko M, Lapish CC, Tosi Z, Hottowy P, Smith WC, Masmanidis SC, Litke AM, Sporns O, Beggs JM (2016) Rich-club organization in effective connectivity among cortical neurons. J Neurosci 36(3):670–684
Peterson BS, Dong ZC (2013) A support-based reconstruction for SENSE MRI. Sensors 13(4):4029–4040
Rubinov M, Sporns O (2010) Complex network measures of brain connectivity: uses and interpretations. NeuroImage 52(3):1059–1069
Salvador R, Suckling J, Coleman M (2005) Neurophysiological architecture of functional magnetic resonance images of human brain. Cereb Cortex 15(9):1332–1342
Stam CJ, Jones BF, Nolte G (2007) Small-world networks and functional connectivity in Alzheimer’s disease. Cereb Cortex 17(1):92–99
Stam CJ, de Haan W, Daffertshofer A, Jones BF, Manshanden I, van Cappellen van Walsum AM, Montez T, Verbunt JPA, de Munck JC, van Dijk BW, Berendse HW, Scheltens P (2009) Graph theoretical analysis of magnetoencephalographic functional connectivity in Alzheimer's disease. Brain 132(Pt1):213–224
Sun JD (2018) Preliminary study on angiosperm genus classification by weight decay and combination of most abundant color index with fractional Fourier entropy. Multimed Tools Appl 77(17):22671–22688
Sun JF, Hong XF, Tong SB (2011) Research progress of complex brain networks-structure, function, calculation and application. J Complex Syst Complexity 7(4):74–90
Tang CS (2018) Multiple sclerosis identification by convolutional neural network with dropout and parametric ReLU. J Comput Sci 28:1–10
Tobia MJ, Hayashi K, Ballard G, Gotlib IH, Waugh CE (2017) Dynamic functional connectivity and individual differences in emotions during social stress. Hum Brain Mapp 38(12):6185–6205
Tzourio-Mazoyer N, Landeau B, Papathanassiou D, Crivello F, Etard O, Delcroix N, Mazoyer B, Joliot M (2002) Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. NeuroImage 15(1):273–289
Wang JH, Zuo X, He Y (2010) Graph-based network analysis of resting-state functional MRI. Front Syst Neurosci 4:16
Wang KC, Wu GB, Hou X, Wei DT, Liu HS, Qiu J (2016) Segmentation and application of functional network from group to individual. Sci Bull 61(27):3022–3035
Wang X, Ren YS, Zhang WS (2017) Multi-task fused lasso method for constructing dynamic functional brain network of resting-state fMRI. J Image Graphics 22(7):0978–0987
Wu LN, Wang SH, Wei G (2010) Color image enhancement based on HVS and PCNN. SCIENCE CHINA Inf Sci 53(10):1963–1976
Yang Z, Zuo XN, Mcmahon KL (2016) Genetic and environmental contributions to functional connectivity architecture of the human brain. Cereb Cortex 26(5):2341–2352
Zhang YD, Wu LN (2008) Improved image filter based on SPCNN. Sci China Ser F Inf Sci 51(12):2115–2125
Zhang YD, Wu LN (2008) Weights optimization of neural network via improved BCO approach. Prog Electromagn Res 83:185–198
Zhang YD, Wu LN (2008) Pattern recognition via PCNN and Tsallis entropy. Sensors 8(11):7518–7529
Zhang YD, Wu LN (2009) Segment-based coding of color images. Sci China F Ed Inf Sci 52(6):914–925
Zhang YD, Wu LN (2011) Optimal multi-level thresholding based on maximum Tsallis entropy via an artificial bee colony approach. Entropy 13(4):841–859
Zhang YD, Wu LN (2012) An MR brain images classifier via principal component analysis and kernel support vector machine. Prog Electromagn Res 130:369–388
Zhang YD, Wu LN, Wei G (2009) A new classifier for polarimetric SAR images. Prog Electromagn Res-PIER 94(4):83–104
Zhang YD, Wu LN, Peterson BS, Dong ZC (2011) A two-level iterative reconstruction method for compressed sensing MRI. J Electromagn Wave Appl 25(8–9):1081–1091
Zhang YD, Phillips P, Wang SH, Ji GL, Yang JQ (2015) Exponential wavelet iterative shrinkage thresholding algorithm for compressed sensing magnetic resonance imaging. Inf Sci 322:115–132
Zhang Y, Zhang H, Chen XB, Lee SW, Shen DG (2017) Hybrid high-order functional connectivity networks using resting-state functional MRI for mild cognitive impairment diagnosis. Sci Rep 7(1):6530
Zhang YD, Pan CC, Chen XQ, Wang F (2018) Abnormal breast identification by nine-layer convolutional neural network with parametric rectified linear unit and rank-based stochastic pooling. J Comput Sci 27:57–68
Zhang YD, Muhammad K, Tang CS (2018) Twelve-layer deep convolutional neural network with stochastic pooling for tea category classification on GPU platform. Multimed Tools Appl 77(17):22821–22839
Zhao XW, Yan JZ, Liang PP (2016) Human brain function partitioning for fMRI data. Chin Sci Bull 61(18):2035–2052
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant No. 51877013, No. 51307010), the Natural Science Foundation of Jiangsu Province (Grant No. BK20181463), the Key Research and Development Plan of Jiangsu Science and Technology Department (Grant No. BE2018638), the Science and Technology Program of Changzhou City (Grant No. CE20185038) and the University Natural Science Research Program of Jiangsu Province (Grant No. 17KJB510003, No. 13KJB510002).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Jiao, Z., Cai, M., Ming, X. et al. Module dividing for brain functional networks by employing betweenness efficiency. Multimed Tools Appl 79, 15253–15271 (2020). https://doi.org/10.1007/s11042-018-7125-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-018-7125-8