Abstract
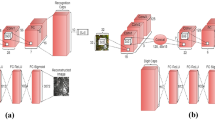
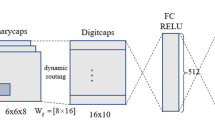
With new architectures providing astonishing performance on many vision tasks, the interest in Convolutional Neural Networks (CNNs) has grown exponentially in the recent past. Such architectures, however, are not problem-free. For instance, one of the many issues is that they require a huge amount of labeled data and are not able to encode pose and deformation information. Capsule Networks (CapsNets) have been recently proposed as a solution to the issues related to CNNs. CapsNet achieved interesting results in images recognition by addressing pose and deformation encoding challenges. Despite their success, CapsNets are still an under-investigated architecture with respect to the more classical CNNs. Following the ideas of CapsNet, we propose to introduce Residual Capsule Network (ResNetCaps) and Dense Capsule Network (DenseNetCaps) to tackle the image recognition problem. With these two architectures, we expand the encoding phase of CapsNet by adding residual convolutional and densely connected convolutional blocks. In addition to this, we investigate the application of feature interaction methods between capsules to promote their cooperation while dealing with complex data. Experiments on four benchmark datasets demonstrate that the proposed approach performs better than existing solutions.







Similar content being viewed by others
Notes
Code will be made available upon acceptance.
References
Akar E, Marques O, Andrews W, Furht B (2019) Cloud-based skin lesion diagnosis system using convolutional neural networks. In: Intelligent computing, pp 982–1000
Akcay S, Kundegorski ME, Willcocks CG, Breckon TP (2018) Using deep convolutional neural network architectures for object classification and detection within x-ray baggage security imagery. IEEE Trans Inf Forens Secur 13 (9):2203–2215
Armenteros JJA, Tsirigos KD, Sønderby CK, Petersen TN, Winther O, Brunak S, von Heijne G, Nielsen H (2019) Signalp 5.0 improves signal peptide predictions using deep neural networks. Nat Biotechnol 37(4):420
Asuntha A, Srinivasan A (2020) Deep learning for lung cancer detection and classification. Multimed Tools Appl:1–32
Bakkouri I, Afdel K (2019) Computer-aided diagnosis (cad) system based on multi-layer feature fusion network for skin lesion recognition in dermoscopy images. Multimed Tools Appl:1–36
Barbuti R, Chessa S, Micheli A, Pucci R (2013) Identification of nesting phase in tortoise populations by neural networks. extended abstract. In: The 50th anniversary convention of the AISB, selected papers, pp 62–65
Bi L, Kim J, Ahn E, Feng D (2017) Automatic skin lesion analysis using large-scale dermoscopy images and deep residual networks. arXiv:1703.04197
Chao H, Dong L, Liu Y, Lu B (2019) Emotion recognition from multiband eeg signals using capsnet. Sensors 19(9):2212
Chessa S, Micheli A, Pucci R, Hunter J, Carroll G, Harcourt R (2017) A comparative analysis of svm and idnn for identifying penguin activities. Appl Artif Intell 31(5-6):453–471
Deliège A, Cioppa A, Van droogenbroeck M (2018) Hitnet: a neural network with capsules embedded in a hit-or-miss layer, extended with hybrid data augmentation and ghost capsules. arXiv:1806.06519
Esteva A, Kuprel B, Novoa RA, Ko J, Swetter SM, Blau HM, Thrun S (2017) Dermatologist-level classification of skin cancer with deep neural networks. Nature 542(7639):115
Esteva A, Robicquet A, Ramsundar B, Kuleshov V, DePristo M, Chou K, Cui C, Corrado G, Thrun S, Dean J (2019) A guide to deep learning in healthcare. Nat Med 25(1):24
Ferentinos KP (2018) Deep learning models for plant disease detection and diagnosis. Comput Electron Agric 145:311–318
Habibzadeh M, Jannesari M, Rezaei Z, Baharvand H, Totonchi M (2018) Automatic white blood cell classification using pre-trained deep learning models: Resnet and inception. In: ICMV 2017, vol 10696, pp 1069612
Han SS, Park GH, Lim W, Kim MS, Im Na J, Park I, Chang SE (2018) Deep neural networks show an equivalent and often superior performance to dermatologists in onychomycosis diagnosis: Automatic construction of onychomycosis datasets by region-based convolutional deep neural network. Plos one 13(1):e0191493
He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. In: CVPR, pp 770–778
Hinrichs A, Vybíral J (2011) Johnson-lindenstrauss lemma for circulant matrices. Random Struct Algorithm 39(3):391–398
Hinton G, Krizhevsky A, Wang SD (2011) Transforming auto-encoders. In: ICANN, pp 44–51
Hinton G, Sabour S, Frosst N (2018) Matrix capsules with em routing
Hou L, Cheng Y, Shazeer N, Parmar N, Li Y, Korfiatis P, Drucker TM, Blezek DJ, Song X (2019) High resolution medical image analysis with spatial partitioning. arXiv:1909.03108
Huang G, Liu Z, Van Der Maaten L, Weinberger KQ (2017) Densely connected convolutional networks. In: CVPR, pp 4700–4708
Huang Y, Cheng Y, Chen D, Lee H, Ngiam J, Le QV, Chen Z (2018) Gpipe: Efficient training of giant neural networks using pipeline parallelism. arXiv:1811.06965
Jiménez J, Skalic M, Martinez-Rosell G, De Fabritiis G (2018) K deep: protein–ligand absolute binding affinity prediction via 3d-convolutional neural networks. J Chem Inf Model 58(2):287–296
Kang MJ, Kang JW (2016) Intrusion detection system using deep neural network for in-vehicle network security. Plos one 11(6):e0155781
Kermany DS, Goldbaum M, Cai W, Valentim CC, Liang H, Baxter SL, McKeown A, Yang G, Wu X, Yan F et al (2018) Identifying medical diagnoses and treatable diseases by image-based deep learning. Cell 172 (5):1122–1131
Kingma DP, Ba J (2014) Adam: A method for stochastic optimization. arXiv:1412.6980
Kosiorek A, Sabour S, Teh YW, Hinton G (2019) Stacked capsule autoencoders. In: Advances in neural information processing systems, pp 15486–15496
Krizhevsky A, Hinton G (2009) Learning multiple layers of features from tiny images. Technical report
LeCun Y, Boser BE, Denker JS, Henderson D, Howard RE, Hubbard WE, Jackel LD (1990) Handwritten digit recognition with a back-propagation network. In: NIPS, pp 396–404
LeCun Y, Bengio Y, Hinton G (2015) Deep learning. Nature 521(7553):436
Lin TY, RoyChowdhury A, Maji S (2015) Bilinear cnn models for fine-grained visual recognition. In: ICCV, pp 1449–1457
Liu W, Wang Z, Liu X, Zeng N, Liu Y, Alsaadi FE (2017) A survey of deep neural network architectures and their applications. Neurocomputing 234:11–26
Liu W, Barsoum E, Owens JD (2018) Object localization with a weakly supervised capsnet. arXiv:1805.07706
Liu JW, Ding XH, Lu RK, Lian YF, Wang D, Luo XL (2019) Multi-view capsule network. In: ICANN, pp 152–165
Martinel N, Micheloni C (2014) Classification of local eigen-dissimilarities for person re-identification. IEEE Signal Process Lett 22(4):455–459
Martinel N, Micheloni C, Foresti GL (2015) The evolution of neural learning systems: a novel architecture combining the strengths of nts, cnns, and elms. IEEE Syst Man Cybern Mag 1(3):17–26
Morota G, Ventura RV, Silva FF, Koyama M, Fernando SC (2018) Machine learning and data mining advance predictive big data analysis in precision animal agriculture. Journal of Animal Science
Nair P, Doshi R, Keselj S (2018) Pushing the limits of capsule networks. Technical note
Pan X, Luo P, Shi J, Tang X (2018) Two at once: Enhancing learning and generalization capacities via ibn-net. In: ECCV, pp 464–479
Pan X, Shen HB (2018) Predicting rna–protein binding sites and motifs through combining local and global deep convolutional neural networks. Bioinformatics 34(20):3427–3436
Parkhi OM, Vedaldi A, Zisserman A, Jawahar CV (2012) Cats and dogs. In: CVPR
Phaye SSR, Sikka A, Dhall A, Bathula D (2018) Dense and diverse capsule networks: Making the capsules learn better. arXiv:1805.04001
Pucci R, Micheloni C, Roberto V, Foresti GL, Martinel N (2019) An exploration of the interaction between capsules with resnetcaps models. In: ICDSC, pp 3:1–3:6
Rajasegaran J, Jayasundara V, Jayasekara S, Jayasekara H, Seneviratne S, Rodrigo R (2019) Deepcaps: Going deeper with capsule networks. In: CVPR, pp 10725–10733
Rakhlin A, Shvets A, Iglovikov V, Kalinin AA (2018) Deep convolutional neural networks for breast cancer histology image analysis. In: ICIAR, pp 737–744
Rubinstein R The cross-entropy method for combinatorial and continuous optimization. In: Methodology and Computing in Applied Probability, vol 1, pp 127–190
Sabour S, Frosst N, Hinton G (2017) Dynamic routing between capsules. In: NIPS, pp 3856–3866
Schmidhuber J (2015) Deep learning in neural networks: an overview. Neural Netw 61:85–117
Szegedy C, Zaremba W, Sutskever I, Bruna J, Erhan D, Goodfellow I, Fergus R (2013) Intriguing properties of neural networks. arXiv:1312.6199
Tabak MA, Norouzzadeh MS, Wolfson DW, Sweeney SJ, VerCauteren KC, Snow NP, Halseth JM, Di Salvo PA, Lewis JS, White MD et al (2019) Machine learning to classify animal species in camera trap images: applications in ecology. Methods Ecol Evol 10(4):585–590
Wang D, Liu Q (2018) An optimization view on dynamic routing between capsules
Wu A, Han Y (2018) Multi-modal circulant fusion for video-to-language and backward. In: IJCAI, vol 3, pp 8
Xian Y, Lampert CH, Schiele B, Akata Z (2018) Zero-shot learning-a comprehensive evaluation of the good, the bad and the ugly. TPAMI
Xu P, Guo S, Miao Q, Li B, Chen X, Fang D (2018) Face detection of golden monkeys via regional color quantization and incremental self-paced curriculum learning. Multimed Tools Appl 77(3):3143–3170
Zhou T, Li Z, Zhang C, Ma H (2019) Classify multi-label images via improved cnn model with adversarial network. Multimed Tools Appl:1–20
Acknowledgments
We gratefully acknowledge the support of NVIDIA Corporation with the donation of the Titan Xp GPU used for this research. Thanks to Patrizia Papalini for proofreading the article.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Pucci, R., Micheloni, C., Foresti, G.L. et al. Deep interactive encoding with capsule networks for image classification. Multimed Tools Appl 79, 32243–32258 (2020). https://doi.org/10.1007/s11042-020-09455-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-020-09455-8