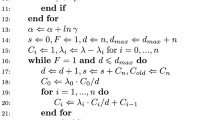Abstract
We develop two prior distributions for probability vectors which, in contrast to the popular Dirichlet distribution, retain sparsity properties in the presence of data. Our models are appropriate for count data with many categories, most of which are expected to have negligible probability. Both models are tractable, allowing for efficient posterior sampling and marginalization. Consequently, they can replace the Dirichlet prior in hierarchical models without sacrificing convenient Gibbs sampling schemes. We derive both models and demonstrate their properties. We then illustrate their use for model-based selection with a hierarchical model in which we infer the active lag from time-series data. Using a squared-error loss, we demonstrate the utility of the models for data simulated from a nearly deterministic dynamical system. We also apply the prior models to an ecological time series of Chinook salmon abundance, demonstrating their ability to extract insights into the lag dependence.









Similar content being viewed by others
References
Agresti, A., Hitchcock, D.B.: Bayesian inference for categorical data analysis. Stat. Methods Appl. 14(3), 297–330 (2005)
Albert, J.H., Gupta, A.K.: Mixtures of Dirichlet distributions and estimation in contingency tables. Ann. Stat. 10, 1261–1268 (1982)
Atchison, J., Shen, S.M.: Logistic-normal distributions: some properties and uses. Biometrika 67(2), 261–272 (1980)
Azat, J.: Grandtab.2016.04.11. California Central Valley Chinook Population Database Report. California Department of Fish and Wildlife. http://www.calfish.org/ProgramsData/Species/CDFWAnadromousResourceAssessment.aspx (2016). Accessed 9 Feb 2019
Berchtold, A., Raftery, A.E.: The mixture transition distribution model for high-order Markov chains and non-Gaussian time series. Stat. Sci. 17, 328–356 (2002)
Besag, J., Mondal, D.: Exact goodness-of-fit tests for Markov chains. Biometrics 69(2), 488–496 (2013)
Bezanson, J., Edelman, A., Karpinski, S., Shah, V.B.: Julia: a fresh approach to numerical computing. SIAM Rev. 59(1), 65–98 (2017)
Bouguila, N., Ziou, D.: Dirichlet-based probability model applied to human skin detection [image skin detection]. In: IEEE International Conference on Acoustics, Speech, and Signal Processing, 2004. Proceedings.(ICASSP’04). IEEE, vol. 5, pp. V–521 (2004)
Connor, R.J., Mosimann, J.E.: Concepts of independence for proportions with a generalization of the Dirichlet distribution. J. Am. Stat. Assoc. 64(325), 194–206 (1969)
Elfadaly, F.G., Garthwaite, P.H.: Eliciting Dirichlet and Gaussian copula prior distributions for multinomial models. Stat. Comput. 27(2), 449–467 (2017)
George, E.I., McCulloch, R.E.: Variable selection via Gibbs sampling. J. Am. Stat. Assoc. 88(423), 881–889 (1993)
George, E.I., McCulloch, R.E.: Approaches for Bayesian variable selection. Stat. Sin. 7, 339–373 (1997)
Good, I.J.: On the application of symmetric Dirichlet distributions and their mixtures to contingency tables. Ann. Stat. 4, 1159–1189 (1976)
Green, P.J.: Reversible jump Markov chain Monte Carlo computation and Bayesian model determination. Biometrika 82, 711–732 (1995)
Hamilton, N.: ggtern: An Extension to ‘ggplot2’, for the Creation of Ternary Diagrams. R package version 2.2.1. https://CRAN.R-project.org/package=ggtern (2017). Accessed 9 Feb 2019
Hare, S.R., Mantua, N.J.: An historical narrative on the Pacific Decadal Oscillation, interdecadal climate variability and ecosystem impacts. Report of a Talk Presented at the 20th NE Pacific Pink and Chum Workshop, Seattle, WA (2001)
Hjort, N.L.: Bayesian approaches to non- and semiparametric density estimation. Preprint Series Statistical Research Report. http://urn.nb.no/URN:NBN:no-51774 (1994). Accessed 9 Feb 2019
Insua, D., Ruggeri, F., Wiper, M.: Bayesian Analysis of Stochastic Process Models, vol. 978. Wiley, New York (2012)
Lochner, R.H.: A generalized Dirichlet distribution in Bayesian life testing. J. R. Stat. Soc. Ser. B Methodol. 37, 103–113 (1975)
Park, T., Casella, G.: The Bayesian lasso. J. Am. Stat. Assoc. 103(482), 681–686 (2008)
Prado, R., West, M.: Time Series: Modeling, Computation, and Inference. CRC Press, Boca Raton (2010)
Quinn, T.J., Deriso, R.B.: Quantitative Fish Dynamics. Oxford University Press, Oxford (1999)
Raftery, A.E.: A model for high-order Markov chains. J. R. Stat. Soc. Ser. B Methodol. 47, 528–539 (1985)
Raftery, A., Tavaré, S.: Estimation and modelling repeated patterns in high order Markov chains with the mixture transition distribution model. Appl. Stat. 43, 179–199 (1994)
Satterthwaite, W.H., Carlson, S.M., Criss, A.: Ocean size and corresponding life history diversity among the four run timings of California Central Valley Chinook salmon. Trans. Am. Fish. Soc. 146(4), 594–610 (2017)
Sethuraman, J.: A constructive definition of Dirichlet priors. Stat. Sin. 4, 639–650 (1994)
Tank, A., Fox, E.B., Shojaie, A.: Granger causality networks for categorical time series (2017). arXiv preprint arXiv:1706.02781
Tibshirani, R.: Regression shrinkage and selection via the lasso. J. R. Stat. Soc. Ser. B Methodol. 58, 267–288 (1996)
Wong, T.T.: Generalized Dirichlet distribution in Bayesian analysis. Appl. Math. Comput. 97(2–3), 165–181 (1998)
Zucchini, W., MacDonald, I.L.: Hidden Markov Models for Time Series: An Introduction Using R, vol. 22. CRC press, Boca Raton (2009)
Acknowledgements
The work of the first and the second author was supported in part by the National Science Foundation under award DMS 1310438. The authors gratefully acknowledge helpful comments from an editor and two anonymous referees.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The work of the first and the second author was supported in part by the National Science Foundation under award DMS 1310438.
Appendices
A SBM correction for \(\delta _k\)
Here we derive correction factor (7) proposed to account for sparsity when using the SBM prior as an extension of a Dirichlet prior. We define a probability \(\theta _k\) to be negligible if the corresponding \(Z_k\) is drawn from the first \(\text {Beta}(1,\eta )\) component, or if \(Z_j\) was drawn from the third \(\text {Beta}(\eta , 1)\) distribution for some \(j < k\). Let W be the total number of nonnegligible probabilities in \(\varvec{\theta }\). We can write \(W = W_1 - W_2\) where \(W_1\) is the minimum k such that \(\xi _k=3\) (\(Z_k\) comes from the large component) or K, whichever is smaller, and \(W_2\) counts the number of times \(\xi _k=1\) (\(Z_k\) comes from the small component) among \(k = 1, \ldots , W_1 - 1\). \(W_1\) comes from a truncated geometric distribution with success probability \(\pi _3\) and has expectation \(\text {E}(W_1) = (1 - (1 - \pi _3)^K)/\pi _3\). Conditional on \(W_1\), \(W_2\) is binomial with \(W_1 - 1\) trials and success probability \(\pi _1\) and thus has conditional expectation \(\text {E}(W_2 \mid W_1) = (W_1 - 1)\pi _1\). Putting these together, we find
Substituting \(\text {E}(W_1)\) and dividing by K yields (7). In the \(\pi _3 = 0\) case, we have \(W_1 = K\) with probability 1, so that \(\text {E}(W_1) = K\).
B Marginal distributions
We report the marginal distributions of observations associated with the Dirichlet, SDM and SBM models for probability vectors. These distributions can be useful for computing Bayes factors in addition to facilitating the MCMC algorithm described in “Appendix C.2.”
Consider a sequence of independent random variables \(\{s_t\} \in \{1, \ldots , K\}^T\) with common distribution \(\varvec{\theta } \equiv (\theta _1, \ldots , \theta _K)\). Given \(\varvec{\theta }\), the probability of the sequence is \(\prod _{t} \theta _{s_t} = \theta _1^{n_1} \cdots \theta _K^{n_K}\) where the sufficient statistics in \(\varvec{n} = (n_1, \ldots , n_K)\) count the occurrences of each category. If the ordering t is not important, the probability is multiplied by the multinomial coefficient \(T! / (n_1! \cdots n_K!)\).
If \(\varvec{\theta }\) follows a Dirichlet distribution with shape parameter vector \(\varvec{\alpha }\), then the marginal (prior predictive) distribution of \(\{s_t\}\) is given by
where \(\text {MVB}(\cdot )\) denotes the multivariate beta function and the integral is defined on the simplex supporting \(\varvec{\theta }\).
If \(\varvec{\theta }\) follows the SDM distribution with parameters \(\varvec{\alpha }\) and \(\beta \), and \(w_k = \prod _{j=1}^K \varGamma (\alpha _j + \beta 1_{(j=k)})\), then the marginal distribution of \(\{s_t\}\) is given by
where \(\varvec{e}_k\) denotes a vector of 0s with a 1 in the kth entry.
Before considering the SBM model, we first obtain the marginal distribution under the generalized Dirichlet (Connor and Mosimann 1969). Working directly with the stick-breaking weights, we have \(p(\varvec{Z}) = \prod _{k=1}^{K-1} \text {Beta}( Z_k ; a_k, b_k )\) and \(p( \{s_t\} \mid \varvec{Z}) = Z_1^{n_1} (1-Z_1)^{\sum _{k=2}^K n_k} \cdots Z_{K-1}^{n_{K-1}} (1-Z_{K-1})^{n_K} \). Putting these together and integrating over \(\varvec{Z}\) results in \(p(\{s_t\}) = \prod _{k=1}^{K-1} g_k(a_k, b_k, \varvec{n}) \) where
with \(a_k^* = a_k + n_k\) and \(b_k^* = b_k + \sum _{j=k+1}^K n_j \, \). Using a similar approach, it can be shown that under the SBM model with parameters \(\varvec{\pi }\), \(\eta \), \(\varvec{\gamma }\) and \(\varvec{\delta }\), we have
C MCMC algorithm details
Following the hierarchical MTD model outlined in (10), the joint posterior distribution of all unknown parameters is given up to proportionality:
1.1 C.1 Original algorithm
MCMC sampling for the original hierarchical MTD structure (Insua et al. 2012) can be achieved entirely with Gibbs updates. This is also the case when substituting in the SDM and SBM priors. A Gibbs sampler cycles through the parameters, drawing updates from the conditional distributions given below.
-
\(\Pr ( \zeta _t = \ell \mid \cdots ) \propto \Pr ( \zeta _t = \ell \mid \varvec{\lambda } ) \, p( s_t \mid \zeta _t = \ell , \varvec{Q} ) = \lambda _\ell \, (\varvec{Q})_{s_{t-\ell }, s_t}\), independently for each \(t \in \{R+1, \ldots , T\}\).
-
\(p(\varvec{\lambda } \mid \cdots ) \propto p(\varvec{\lambda })\, \prod _t p(\zeta _t \mid \varvec{\lambda }) = \text {Dir}( \varvec{\lambda }; \varvec{\alpha }_\lambda ) \, \prod _t \lambda _{\zeta _t} \), a standard Dirichlet/multinomial update using the counts of \(\zeta _t\) in each of \(\{1, \ldots , R\}\). In the case of a SDM prior for \(\varvec{\lambda }\), this becomes the standard SDM update given in Sect. 2.1. In the case of a SBM prior for \(\varvec{\lambda }\), this becomes the standard SBM update given in Sect. 2.2 which draws updated latent stick-breaking weights and constructs \(\varvec{\lambda }\).
-
\(p( \varvec{Q}_{k,\cdot } \mid \cdots ) \propto p(\varvec{Q}_{k,\cdot }) \prod _{t : s_{t-\zeta _t} = k} p(s_t \mid \zeta _t, \varvec{Q}_{k,\cdot }) = \text {Dir}( \varvec{Q}_{k, \cdot }; \varvec{\alpha }_Q ) \, \prod _{t : s_{t-\zeta _t} = k} (\varvec{Q})_{k, s_t} \), which is again a standard Dirichlet/multinomial update using transition counts. In the case of a SBM prior for the kth row of \(\varvec{Q}\), this becomes the standard SBM update given in Sect. 2.2 which draws updated latent stick-breaking weights and constructs the row of \(\varvec{Q}\).
This Gibbs sampler can be modified to include an update for \(\varvec{\pi }_k\) together with the update for the kth row of \(\varvec{Q}\) since the only component of the posterior in (15) dependent on \(\varvec{\pi }_k\) is \(p(\varvec{Q}_{k,\cdot })\). A Dirichlet prior for \(\varvec{\pi }_k\) results in a Dirichlet full conditional, using the counts of latent \(\xi \) variables drawn using (6). Updates for any other hyperparameters would require an alternate sampling scheme.
1.2 C.2 Modified algorithm
Sampling \(\{\zeta _t \}\) conditional on \(\varvec{Q}\), followed by \(\varvec{Q}\) conditional on \(\{\zeta _t\}\) results in a chain that tends to get stuck. To improve mixing, we instead integrate \(\varvec{Q}\) out of joint posterior (15) and conduct Gibbs sampling between \(\{\zeta _t \}\) and \(\varvec{\lambda }\). At each iteration, it is then straightforward to draw \(\varvec{Q}\) from the conditional distribution given in 1.
Let \(\varvec{N}\) be a \(K\times K\) matrix of transition counts for which the \((k_1, k_2)\) entry is the cardinality of \(\{ t : s_{t - \zeta _t} = k_1 \ \text {and} \ s_t = k_2 \}\). Integrating \(\varvec{Q}\) from (15) yields \( p(\{\zeta _t\}, \varvec{\lambda } \mid \{s_t\}) \propto p(\varvec{\lambda }) \, \prod _t [ p(\zeta _t \mid \varvec{\lambda }) \, p(s_t \mid \zeta _t) ] \) which differs from the original only in that
where the \(h(\cdot , \phi )\) takes the form of (12) if the rows of \(\varvec{Q}\) are independent Dirichlet, (13) if the rows of \(\varvec{Q}\) are independent SDM, and (14) if the rows of \(\varvec{Q}\) are independent SBM; and \(\phi \) refers to generic hyperparameters appropriate for the choice of prior.
The modified algorithm then proceeds with the standard update for \(\varvec{\lambda }\) given in C.1. Each \(\zeta _t \) is then updated individually using its full conditional involving \(\varvec{\lambda }\) and (16) by modifying \(\varvec{N}\) to reflect each possible value of \(\zeta _t \in \{1, \ldots , R\}\). Unnecessary computation can be avoided by noting redundancies in the denominators of (12) and (13), and computing (16) only over values of k such that \(s_{t-\ell } = k\) for \(\ell = 1, \ldots , R\).
Allowing inference for \(\{\varvec{\pi }_k\}\) when utilizing SBM priors for \(\varvec{Q}\) is no more complicated than in the original algorithm. Simply draw \(\varvec{\pi }_k\) when sampling from the conditional for the kth row of \(\varvec{Q}\). Note however that integrating over \(\varvec{Q}\) leaves the update for \(\varvec{\lambda }\) conditional on \(\{\varvec{\pi }_k\}\). Nevertheless, the result is still a valid Gibbs sampler which in practice produces acceptable mixing.
Rights and permissions
About this article
Cite this article
Heiner, M., Kottas, A. & Munch, S. Structured priors for sparse probability vectors with application to model selection in Markov chains. Stat Comput 29, 1077–1093 (2019). https://doi.org/10.1007/s11222-019-09856-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11222-019-09856-2