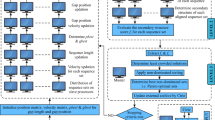
Abstract
Protein structure prediction (PSP) is an open problem with many useful applications in disciplines such as medicine, biology and biochemistry. As this problem presents a vast search space and the analysis of each protein structure requires a significant amount of computing time, it is necessary to take advantage of high-performance parallel computing platforms as well as to define efficient search procedures in the space of possible protein conformations. In this paper we compare two parallel procedures for PSP which are based on different multi-objective optimization approaches, i.e. PAES (Knowles and Corne in Proc. Congr. Evol. Comput. 1:98–105, 1999) and NSGA2 (Deb et al. in IEEE Trans. Evol. Comput. 6:182–197, 2002). Although both procedures include techniques to take advantage of known protein structures and strategies to simplify the search space through the so-called rotamer library and adaptive mutation operators, they present different profiles with respect to their implicit parallelism.
Similar content being viewed by others
References
Knowles J, Corne D (1999) The Pareto archived evolution strategy: a new baseline algorithm for Pareto multiobjective optimisation. Proc Congr Evol Comput 1:98–105
Deb K, Pratap A, Agarwal S, Meyarivan T (2002) A fast and elitist multiobjective genetic algorithm: NSGA-II. IEEE Trans Evol Comput 6:182–197
Zhang Y (2008) Progress and challenges in protein structure prediction. Curr Opin Struct Biol 18:342–348
Cutello V, Narcisi G, Nicosia G (2006) A multi-objective evolutionary approach to the protein structure prediction problem. J R Soc Interface 3:139–151
Anfinsen C (1973) Principles that govern the folding of protein chains. Science 181:223–230
Zhang Y, Skolnick J (2005) The protein structure prediction problem could be solved using the current PDB library. Proc Natl Acad Sci USA 102:1029–1034
Handl J, Kell D, Knowles J (2007) Multiobjective optimization in bioinformatics and computational biology. IEEE/ACM Trans Comput Biol Bioinform (TCBB) 4:279–292
Rychlewski L, Jaroszewski L, Li W, Godzik A (2000) Comparison of sequence profiles. Strategies for structural predictions using sequence information. Protein Science 9:232–241
Raman S et al (2008) Advances in Rosetta protein structure prediction on massively parallel systems. IBM J Res Dev 52:7–17
Bradley P, Misura K, Baker D (2005) Toward high-resolution de novo structure prediction for small proteins. Science 309:1868–1871
Taufer M, An C, Kerstens A, Brooks C (2006) Predictor@home: a ‘protein structure prediction supercomputer’ based on global computing. IEEE Trans Parallel Distrib Syst 17:786–796
Anderson D (2004) Boinc: a system for public-resource computing and storage. In: Proc Fifth IEEE/ACM Int’l. Workshop Grid Computing
Coello CAC, Lamont GB, Veldhuizen DAV (2007) Evolutionary algorithms for solving multi-objective problems. SpringerLink e-books
Veldhuizen DV, Zidallis J, Lamont G (2003) Considerations in engineering parallel multiobjective evolutionary algorithms. IEEE Trans Evol Comput 7:144–173
Tantar A-A, Melab N, Talbi E-G, Parent B, Horvath D (2007) A parallel hybrid genetic algorithm for protein structure prediction on the computational grid. Future Gener Comput Syst 23:398–409
Day R, Zydallis J, Lamont G (2002) Solving the protein structure prediction problem through a multiobjective genetic algorithm. Nanotech 2:32–35
Rohl C, Strauss C, Misura K, Baker D (2004) Protein structure prediction using Rosetta. Methods Enzymol 383:66–93
Wu S, Skolnick J, Zhang Y (2007) Ab initio modelling of small proteins by iterative TASSER simulations. BMC Biol 5:17
Helles G (2007) A comparative study of the reported performance of ab initio protein structure prediction algorithms. J R Soc Interface 5:387–396
Dunbrack R (2002) Rotamer libraries in the 21st century. Curr Opin Struct Biol 12:431–440
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Calvo, J.C., Ortega, J. & Anguita, M. Comparison of parallel multi-objective approaches to protein structure prediction. J Supercomput 58, 253–260 (2011). https://doi.org/10.1007/s11227-009-0368-4
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11227-009-0368-4