Abstract
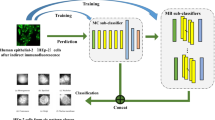
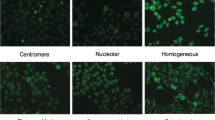
Identifying human epithelial-2 (HEp-2) cells in indirect immune fluorescence (IIF) is the most commonly used method for the diagnosis of autoimmune diseases. Although supervised deep learning networks have made remarkable progress on HEp-2 cell staining pattern classification, the high-performance relies on a large amount of labeled training data. Unfortunately, large-scale labeled datasets are scarce due to the expensive costs of labeling medical images. Therefore, we propose an unsupervised domain adaption (UDA) model, namely MDA-MPCD, to classify unlabeled HEp-2 cell samples. The proposed model involves two major aspects: (a) multi-context domain adaption (MDA) generator and (b) maximum partial classifier discrepancy (MPCD) architecture. The MDA generator can extract multi-context features from complex cell images while providing more comprehensive and diverse information for the classifier. The MPCD architecture, utilizing the mapping variation of feature transfer, focuses on the discrepancy from the cross-domain gap by separating the activations in the classifier. The proposed model dominates all comparison methods during evaluation, achieving 85.35% and 96.08% mean accuracy on two UDA tasks, respectively. Furthermore, the model is demonstrated to adapt from rich label domain to unlabeled domain by detailed ablation experiments and visualization results. The proposed MDA-MPCD has potential as a clinical scheme, enabling effective and accurate classification of HEp-2 cell staining pattern without labeling the target data.









Similar content being viewed by others
Notes
Download link: https://hep2.unisa.it/dbtools.html.
Download link: https://mivia.unisa.it/contest-hep-2/.
References
Rahman S, Wang L, Sun C, Zhou L (2020) Deep learning based HEp-2 image classification: a comprehensive review. Med Image Anal. https://doi.org/10.1016/j.media.2020.101764
Kavanaugh A, Tomar R, Reveille J, Solomon DH, Homburger HA (2000) Guidelines for clinical use of the antinuclear antibody test and tests for specific autoantibodies to nuclear antigens. Arch Pathol Lab Med 124:71–81. https://doi.org/10.5858/2000-124-0071-GFCUOT
Bayramoglu N, Kannala J, Heikkilä J (2015) Human epithelial type 2 cell classification with convolutional neural networks. In: 2015 IEEE 15th International Conferences Bioinformatics-Bioengineering BIBE, pp 0–5. https://doi.org/10.1109/BIBE.2015.7367705
Rodrigues LF, Naldi MC, Mari JF (2020) Comparing convolutional neural networks and preprocessing techniques for HEp-2 cell classification in immunofluorescence images. Comput Biol Med 116:103542. https://doi.org/10.1016/j.compbiomed.2019.103542
Ahn E, Kumar A, Fulham M, Feng D, Kim J (2020) Unsupervised domain adaptation to classify medical images using zero-bias convolutional auto-encoders and context-based feature augmentation. IEEE Trans Med Imaging 39:2385–2394. https://doi.org/10.1109/TMI.2020.2971258
Phan HTH, Kumar A, Kim J, Feng D (2016) Transfer learning of a convolutional neural network for HEp-2 cell image classification. Proc Int Symp Biomed Imaging 2:1208–1211. https://doi.org/10.1109/ISBI.2016.7493483
Li H, Zheng W-S, Zhang J (2016) Deep CNNs for HEp-2 cells classification : a cross-specimen analysis. http://arxiv.org/abs/1604.05816
Shen L, Jia X, Li Y (2018) Deep cross residual network for HEp-2 cell staining pattern classification. Pattern Recognit 82:68–78. https://doi.org/10.1016/j.patcog.2018.05.005
Lei H, Han T, Zhou F, Yu Z, Qin J, Elazab A, Lei B (2018) A deeply supervised residual network for HEp-2 cell classification via cross-modal transfer learning. Pattern Recognit 79:290–302. https://doi.org/10.1016/j.patcog.2018.02.006
Foggia P, Percannella G, Soda P, Vento M (2013) Benchmarking HEp-2 cells classification methods. IEEE Trans Med Imaging 32:1878–1889. https://doi.org/10.1109/TMI.2013.2268163
Wu S, Wang F, Huang J, Yin B, Huang M, Wei W, Zhang M, Ouyang R (2019) Evaluation of the automated indirect immunofluorescence test for anti-dsDNA antibodies. Clin Chim Acta 498:143–147. https://doi.org/10.1016/j.cca.2019.08.018
Hobson P, Lovell BC, Percannella G, Vento M, Wiliem A (2015) Benchmarking human epithelial type 2 interphase cells classification methods on a very large dataset. Artif Intell Med 65:239–250. https://doi.org/10.1016/j.artmed.2015.08.001
Rigon A, Soda P, Zennaro D, Iannello G, Afeltra A (2007) Indirect immunofluorescence in autoimmune diseases: assessment of digital images for diagnostic purpose. Cytom Part B Clin Cytom 72:472–477. https://doi.org/10.1002/cyto.b.20356
Hu B, Tang Y, Chang EIC, Fan Y, Lai M, Xu Y (2019) Unsupervised learning for cell-level visual representation in histopathology images with generative adversarial networks. IEEE J Biomed Heal Inform 23:1316–1328. https://doi.org/10.1109/JBHI.2018.2852639
Zhang Y, Niu S, Qiu Z, Wei Y, Zhao P, Yao J, Huang J, Wu Q, Tan M (2020) COVID-DA: deep domain adaptation from typical pneumonia to COVID-19, vol XX, pp 1–8. http://arxiv.org/abs/2005.01577
Guan H, Liu M (2021) Domain adaptation for medical image analysis: a survey. Science. 5:1–15
Bousmalis K, Trigeorgis G, Silberman N, Krishnan D, Erhan D (2016) Domain separation networks. Adv Neural Inf Process Syst 2:343–351
Tzeng E, Hoffman J, Saenko K, Darrell T (2017) Adversarial discriminative domain adaptation. In: 2017 IEEE Conference Computing Vision and Pattern Recognition, IEEE, pp 2962–2971. https://doi.org/10.1109/CVPR.2017.316
Laradji IH, Babanezhad R (2020) M-ADDA: unsupervised domain adaptation with deep metric learning. Domain Adapt Vis Underst 2:17–31. https://doi.org/10.1007/978-3-030-30671-7_2
Saito K, Watanabe K, Ushiku Y, Harada T (2018) Maximum classifier discrepancy for unsupervised domain adaptation. In: Proceedings of IEEE Computer Society Conference on Computer Vision and Pattern Recognition, pp 3723–3732. https://doi.org/10.1109/CVPR.2018.00392
Lovell BC, Percannella G, Vento M, Wiliem A (2014) Performance evaluation of indirect immunofluorescence image. Anal Syst 2:1–25
Yu F, Koltun V (2016) Multi-scale context aggregation by dilated convolutions. In: 4th International Conference on Learning Representations, ICLR 2016-Conference Track Proceedings
He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. In: Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition. 2016-Decem, pp 770–778. https://doi.org/10.1109/CVPR.2016.90
Gao Z, Wang L, Zhou L, Zhang J (2017) HEp-2 cell image classification with deep convolutional neural networks. IEEE J Biomed Heal Inform 21:416–428. https://doi.org/10.1109/JBHI.2016.2526603
Li Y, Shen L (2019) HEp-Net: a smaller and better deep-learning network for HEp-2 cell classification. Comput Methods Biomech Biomed Eng Imaging Vis 7:266–272. https://doi.org/10.1080/21681163.2018.1449140
Li Y, Shen L, Yu S (2017) HEp-2 specimen image segmentation and classification using very deep fully convolutional network. IEEE Trans Med Imaging 36:1561–1572. https://doi.org/10.1109/TMI.2017.2672702
Pan SJ, Yang Q (2010) A survey on transfer learning. IEEE Trans Knowl Data Eng 22:1345–1359. https://doi.org/10.1109/TKDE.2009.191
Long M, Zhu H, Wang J, Jordan MI (2017) Deep transfer learning with joint adaptation networks. 34th Int Conf Mach Learn ICML 2017(5):3470–3479
Zhuang F, Qi Z, Duan K, Xi D, Zhu Y, Zhu H, Xiong H, He Q (2021) A comprehensive survey on transfer learning. Proc IEEE 109:43–76. https://doi.org/10.1109/JPROC.2020.3004555
Ohata EF, Chagas JVS, Bezerra GM, Hassan MM, Albuquerque VHC, Filho PPR (2021) A novel transfer learning approach for the classification of histological images of colorectal cancer. J Supercomput 77:9494–9519. https://doi.org/10.1007/s11227-020-03575-6
Vununu C, Lee S-H, Kwon K-R (2021) A classification method for the cellular images based on active learning and cross-modal transfer learning. Sensors 21:1469. https://doi.org/10.3390/s21041469
Zhang Y, Wei Y, Wu Q, Zhao P, Niu S, Huang J, Tan M (2020) Collaborative unsupervised domain adaptation for medical image diagnosis. IEEE Trans Image Process 29:7834–7844. https://doi.org/10.1109/TIP.2020.3006377
Zhuang J, Chen Z, Zhang J, Zhang D, Cai Z (2019) Domain adaptation for retinal vessel segmentation using asymmetrical maximum classifier discrepancy. ACM Int Conf Proc Ser. https://doi.org/10.1145/3321408.3322627
Ronneberger O, Fischer P, Brox T (2015) U-Net: convolutional networks for biomedical image segmentation. In: IEEE Access, pp 234–241. https://doi.org/10.1007/978-3-319-24574-4_28
Yosinski J, Clune J, Bengio Y, Lipson H (2014) How transferable are features in deep neural networks? Adv Neural Inf Process Syst 4:3320–3328
Xu R, Li G, Yang J, Lin L (2019) Larger norm more transferable: an adaptive feature norm approach for unsupervised domain adaptation. In: Proceedings of the IEEE International Conference on Computer Vision, pp 1426–1435. https://doi.org/10.1109/ICCV.2019.00151
Sokolova M, Lapalme G (2009) A systematic analysis of performance measures for classification tasks. Inf Process Manag 45:427–437. https://doi.org/10.1016/j.ipm.2009.03.002
LeCun Y, Bottou L, Bengio Y, Haffner P (1998) Gradient-based learning applied to document recognition. Proc IEEE 86:2278–2323. https://doi.org/10.1109/5.726791
Simonyan K, Zisserman A (2015) Very deep convolutional networks for large-scale image recognition. In: 3rd International Conference on Learning Representations ICLR 2015-Conference Track Proceedings, pp 1–14
Szegedy C, Vanhoucke V, Ioffe S, Shlens J, Wojna Z (2016) Rethinking the inception architecture for computer vision. In: Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition 2016-Decem, pp 2818–2826. https://doi.org/10.1109/CVPR.2016.308
Nguyen LD, Gao R, Lin D, Lin Z (2019) Biomedical image classification based on a feature concatenation and ensemble of deep CNNs. J Ambient Intell Humaniz Comput. https://doi.org/10.1007/s12652-019-01276-4
Acknowledgements
This research is funded by the Fundamental Research Funds for the Central Universities under Grant No. N181706001, N2017009, N2017008, N182608003 and N181703005, National Natural Science Foundation of China under Grant No.61902057 and Joint Fund of Science and Technology Department of Liaoning Province and State Key Laboratory of Robotics China under Grant No. 2020-KF-12-11.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Zhao, H., Ren, T., Wang, C. et al. Multi-context unsupervised domain adaption for HEp-2 cell classification using maximum partial classifier discrepancy. J Supercomput 78, 14362–14380 (2022). https://doi.org/10.1007/s11227-022-04452-0
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11227-022-04452-0