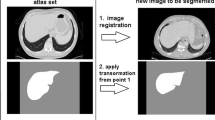
Abstract
Purpose
This paper addresses joint optimization for segmentation and shape priors, including translation, to overcome inter-subject variability in the location of an organ. Because a simple extension of the previous exact optimization method is too computationally complex, we propose a fast approximation for optimization. The effectiveness of the proposed approximation is validated in the context of gallbladder segmentation from a non-contrast computed tomography (CT) volume.
Methods
After spatial standardization and estimation of the posterior probability of the target organ, simultaneous optimization of the segmentation, shape, and location priors is performed using a branch-and-bound method. Fast approximation is achieved by combining sampling in the eigenshape space to reduce the number of shape priors and an efficient computational technique for evaluating the lower bound.
Results
Performance was evaluated using threefold cross-validation of 27 CT volumes. Optimization in terms of translation of the shape prior significantly improved segmentation performance. The proposed method achieved a result of 0.623 on the Jaccard index in gallbladder segmentation, which is comparable to that of state-of-the-art methods. The computational efficiency of the algorithm is confirmed to be good enough to allow execution on a personal computer.
Conclusions
Joint optimization of the segmentation, shape, and location priors was proposed, and it proved to be effective in gallbladder segmentation with high computational efficiency.














Similar content being viewed by others
References
Shimizu A, Ohno R, Ikegami T, Kobatake H, Nawano S, Smutek D (2007) Segmentation of multiple organs in non-contrast 3D abdominal CT images. Int J Comput Assist Radiol Surg 2(3–4):135–142. doi:10.1007/s11548-007-0135-z
Rueckert D, Linguraru M, Pura J, Pamulapati V, Summers R (2012) Statistical 4D graphs for multi-organ abdominal segmentation from multiphase CT. Med Image Anal 16(4):904–914
Wolz R, Chu C, Misawa K, Fujiwara M, Mori K, Rueckert D (2013) Automated abdominal multi-organ segmentation with subject-specific atlas generation. IEEE Trans Med Imaging 32(9):1723–1730. doi:10.1109/TMI.2013.2265805
Li C, Wang X, Li J, Eberl S, Fulham M, Yin Y, Feng D (2013) Joint probabilistic model of shape and intensity for multiple abdominal organ segmentation from volumetric CT images. IEEE J Biomed Health Inf 17(1):92–102
Oshima R, Saito A, Shimizu A, Nawano S (2015) Improvement of abdominal multi-organ segmentation using a relaxed conditional hierarchical statistical shape model. In: Proc. of the international forum on medical imaging in Asia (IFMIA), OS-2
Cerrolaza J, Reyes M, Summers R, GonzAlez-Ballester M, Linguraru M (2015) Automatic multi-resolution shape modeling of multi-organ structures. Med Image Anal 25(1):11–21
Tomoshige S, Oost E, Shimizu A, Watanabe H, Nawano S (2014) A conditional statistical shape model with integrated error estimation of the conditions: application to liver segmentation in non-contrast CT images. Med Image Anal 18(1):130–143. doi:10.1016/j.media.2013.10.003
Okada T, Linguraru MG, Hori M, Summers RM, Tomiyama N, Sato Y (2015) Abdominal multi-organ segmentation from CT images using conditional shape–location and unsupervised intensity priors. Med Image Anal 26(1):1–18. doi:10.1016/j.media.2015.06.009
Linguraru MG, Pura JA, Pamulapati V, Summers RM (2012) Statistical 4D graphs for multi-organ abdominal segmentation from multiphase CT. Med Image Anal 16(4):904–914. doi:10.1016/j.media.2012.02.001
Kass M, Witkin A, Terzopoulos D (1988) Snakes: active contour models. Int J Comput Vis 1(4):321–331. doi:10.1007/BF00133570
Malladi R, Sethian JA, Vemuri BC (1995) Shape modeling with front propagation: a level set approach. IEEE Trans Pattern Anal Mach Intell 17(2):158–175. doi:10.1109/34.368173
Boykov Y, Veksler O, Zabih R (2001) Fast approximate energy minimization via graph cuts. IEEE Trans Pattern Anal Mach Intell 23(11):1222–1239. doi:10.1109/34.969114
Heimann T, Meinzer HP (2009) Statistical shape models for 3D medical image segmentation: a review. Med Image Anal 13(4):543–563. doi:10.1016/j.media.2009.05.004
Cootes T, Taylor C, Cooper D, Graham J (1995) Active shape models-their training and application. Comput Vis Image Underst 61:38–59. doi:10.1006/cviu.1995.1004
Cremers D, Rousson M, Deriche R (2007) A review of statistical approaches to level set segmentation: integrating color, texture, motion and shape. Int J Comput Vis 72(2):195–215. doi:10.1007/s11263-006-8711-1
Kohli P, Rihan J, Bray M, Torr PH (2008) Simultaneous segmentation and pose estimation of humans using dynamic graph cuts. Int J Comput Vis 79(3):285–298
Chen X, Udupa JK, Alavi A, Torigian DA (2013) GC–ASM: synergistic integration of graph-cut and active shape model strategies for medical image segmentation. Comput Vis Image Underst 117(5):513–524
Besbes A, Komodakis N, Langs G, Paragios N (2009) Shape priors and discrete MRFs for knowledge-based segmentation. In: Proc. CVPR, pp 1295–1302
Xiang B, Deux JF, Rahmouni A, Paragios N (2013) Joint model-pixel segmentation with pose-invariant deformable graph-priors. In: Proc. MICCAI, LNCS, vol 8151. Springer, Berlin, pp 267–274
Lempitsky V, Blake A, Rother C (2012) Branch-and-mincut: global optimization for image segmentation with high-level priors. J Math Imaging Vis 44(3):315–329
Saito A, Nawano S, Shimizu A (2016) Joint optimization of segmentation and shape prior from level-set-based statistical shape model, and its application to the automated segmentation of abdominal organs. Med Image Anal 28:46–65. doi:10.1016/j.media.2015.11.003
Saito A, Nawano S, Shimizu A (2014) Proposal of an algorithm for simultaneous optimization of segmentation and a shape prior and its application to pancreas segmentation. IEICE tech rep 114(200):1–5
Shimizu A, Kubo M, Furukawa D, Kobatake H, Nawano S (2008) Abdomen standardization for multi-organ segmentation of CT volumes. Int J Comput Assist Radiol Surg 3(supp. 1):s195–s196
Shimizu A, Sakurai H, Kobatake H, Nawano S, Smutek D (2007) Improvement of a multi-organ extraction algorithm in an abdominal CAD system based on features in neighboring regions. Int J Comput Assist Radiol Surg 2(suppl 1):S386–S388. doi:10.1007/s11548-007-0108-2
Uchida Y, Shimizu A, Kobatake H, Nawano S, Shinozaki K (2010) A comparative study of statistical shape models of the pancreas. Int J Comput Assist Radiol Surg 5(suppl 1):S385–S387. doi:10.1007/s11548-010-0469-9
Boomgaard RVD, Balen RV (1992) Methods for fast morphological image transforms using bitmapped binary images. CVGIP: Graph Models Image Process 54(3):252–258
Kohli P, Torr PH (2007) Dynamic graph cuts for efficient inference in Markov random fields. IEEE Trans Pattern Anal Mach Intell 29(12):2079–2088
VISCERAL project. http://www.visceral.eu/. Last accessed 15 Aug 2016
Komodakis K, Xiang B, Paragios N (2015) A framework for efficient structured max-margin learning of high-order MRF models. IEEE Trans Pattern Anal Mach Intell 37(7):1425–1441
Acknowledgements
The authors wish to acknowledge the assistance provided by Dr. Hanaoka Shouhei of Tokyo University for the helpful insight provided from a medical viewpoint. This study was supported in part by a Grant-in-Aid for Scientific Research in Innovative Areas from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (26108002).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
All procedures performed in studies involving human participants were performed in accordance with the ethical standards of the institutional and/or national research committee and with the 1975 Helsinki declaration, as revised in 2008(5).
Informed consent
Formal consent is not required in this type of study.
Appendix
Appendix
The lower bound defined in Eq. (12) satisfies the three properties required for the branch-and-bound method.
- Monotonicity :
-
The inequality \(\textit{LB}(\varTheta _\textit{ch}) \ge \textit{LB}(\varTheta )\) holds for any nested domains \(\varTheta _\textit{ch}\subset \varTheta \). See Corollary 1 with the proof below.
- Tightness :
-
The bound is tight for a singleton node, i.e., \(\textit{LB}(\{\varvec{\theta }\}) = f(\varvec{\theta })\) is satisfied for any \(\varvec{\theta }\in \mathbb {R}^d\times \mathbb {Z}^3\).
- Computability :
-
Evaluation of the lower bound in Eq. (12) \(\varTheta =S\times T\) requires taking the minimum of \(F^i(\varvec{\theta })\) and \(B^i(\varvec{\theta })\) in terms of \(\varvec{\theta }\in \varTheta \), and operation in terms of \(\varvec{\theta }\in \varTheta \), as well as taking the outer minimum in terms of \(\mathbf {x}\in \mathcal {L}^N\). As described in the section “calculation of the lower bound”, the computational complexity of the former minimum operation over \(\varvec{\theta }\in \varTheta \) is \(O(dN+ \log |T|)\). The latter minimum operation equals the minimization of a submodular quadratic pseudo-Boolean function. Theoretical complexity is the low-order polynomial of \(N\) time, if it is solved using s–t mincut [12]. Therefore, computability is satisfied in our study.
Corollary 1
For any nested domains \(\varTheta _\textit{ch}\subset \varTheta \), the inequality \(\textit{LB}(\varTheta _\textit{ch}) \ge \textit{LB}(\varTheta )\) holds.
Proof
Let us denote \(A(\mathbf {x}, \varTheta )\) as the expression within the outer minimum of Eq. (12), i.e.,
Assume \(\varTheta _\textit{ch}\subset \varTheta \). Then, for any fixed \(\mathbf {x}\), for all pixels \(i\in \mathcal {V}\), the following inequalities hold:
This is because \(\varTheta _\textit{ch}\) is a subset of \(\varTheta \) and \(x_i\), and \((1-x_i)\) are nonnegative (\(\because x_i\in \{0, 1\}\)). By summing the inequalities Eqs. (25) and (26) over all pixels (\(i\in \mathcal {V}\)), and adding \(\sum _{(i,j)\in \mathcal {E}}{P^{ij} \cdot \left| x_i- x_j\right| }\) to both sides, which are constant with respect to \(\varvec{\theta }\), we obtain
i.e., monotonicity holds for any constant value of \(\mathbf {x}\). We define \(\mathbf {x}^*_\textit{ch}\) and \(\mathbf {x}^*\) as the segmentation labels obtained by \(\mathbf {x}^*_\textit{ch} = \mathop {\mathrm{arg~ min}}\limits _{\mathbf {x}\in \mathcal {L}^N}A(\mathbf {x}, \varTheta _\textit{ch})\) and \(\mathbf {x}^* = \mathop {\mathrm{arg~ min}}\limits _{\mathbf {x}\in \mathcal {L}^N}A(\mathbf {x}, \varTheta )\), respectively. Then, from the monotonicity in Eq. (27) and the definition of \(\mathbf {x}^*\), we obtain
\(\square \)
Rights and permissions
About this article
Cite this article
Saito, A., Nawano, S. & Shimizu, A. Fast approximation for joint optimization of segmentation, shape, and location priors, and its application in gallbladder segmentation. Int J CARS 12, 743–756 (2017). https://doi.org/10.1007/s11548-017-1571-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11548-017-1571-z