Abstract
Purpose
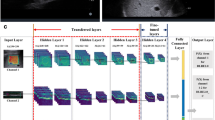
The aim of this study was to explore the application of five-class deep residual network models based on plain CT images and clinical features for the precise staging of liver fibrosis.
Methods
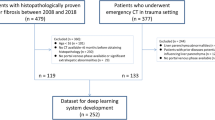
This retrospective clinical study included 347 patients who underwent liver CT, with pathological staging of liver fibrosis as the gold standard. We established three ResNet models to stage liver fibrosis. The output diagnosis labels of models were 0, 1, 2, 3 and 4, which correspond to F0, F1, F2, F3, and F4 stages. Confusion matrices were used to evaluate the performances of models to precisely stage liver fibrosis. The performance for diagnosing cirrhosis (F4), advanced fibrosis (≥ F3) and significant fibrosis (≥ F2) of models was evaluated with receiver operating characteristic (ROC) analyses.
Results
The kappa coefficients of the five-class ResNet model (based on plain CT images), the five-class ResNet clinical model (based on clinical features), and the five-class mixed ResNet model (based on plain CT images and clinical features) for precise staging liver fibrosis were 0.566, 0.306, and 0.63, respectively. The recall rates and precision rates for F0, F1, F2, and F3 of three models were lower than 60%. The ROC AUC values of the five-class ResNet model, the five-class ResNet clinical model, and the five-class mixed ResNet model for diagnosing cirrhosis, advanced fibrosis, and significant fibrosis were 0.95, 0.88, and 0.82, 0.80, 0.72, and 0.70, 0.95, 0.90, and 0.83, respectively.
Conclusions
The five-class ResNet models are of high value in the diagnosis of liver cirrhosis, advanced liver fibrosis, and significant liver fibrosis. However, for the precise staging of liver fibrosis, the models cannot accurately distinguish other liver fibrosis stages except F4. Plain CT images combined with clinical features have the potential to improve the performance of the ResNet models in diagnosing liver fibrosis.







Similar content being viewed by others
References
Lambin P, Rios-Velazquez E, Leijenaar R, Carvalho S, van Stiphout RG, Granton P, Zegers CM, Gillies R, Boellard R, Dekker A, Aerts HJ (2012) Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer 48(4):441–446
Yasaka K, Akai H, Kunimatsu A, Abe O, Kiryu S (2018) Deep learning for staging liver fibrosis on CT: a pilot study. Eur Radiol 28(11):4578–4585
Yasaka K, Akai H, Kunimatsu A, Abe O, Kiryu S (2018) Liver fibrosis: deep convolutional neural network for staging by using gadoxetic acid-enhanced hepatobiliary phase MR images. Radiology 287(1):146–155
Park HJ, Park B, Lee SS (2020) Radiomics and deep learning: hepatic applications. Korean J Radiol 21(4):387–401
Treacher A, Beauchamp D, Quadri B, Fetzer D, Vij A, Yokoo T, Montillo A (2019) Deep learning convolutional neural networks for the estimation of liver fibrosis severity from ultrasound texture. Proc SPIE Int Soc Opt Eng 10950:109503E
Yu Y, Wang J, Ng CW, Ma Y, Mo S, Fong ELS, Xing J, Song Z, Xie Y, Si K, Wee A, Welsch RE, So PTC, Yu H (2018) Deep learning enables automated scoring of liver fibrosis stages. Sci Rep 8(1):16016
Bedossa P, Poynard T (1996) An algorithm for the grading of activity in chronic hepatitis C. The METAVIR Cooperative Study Group. Hepatology 24(2):289–293
Lee JH, Joo I, Kang TW, Paik YH, Sinn DH, Ha SY, Kim K, Choi C, Lee G, Yi J, Bang WC (2020) Deep learning with ultrasonography: automated classification of liver fibrosis using a deep convolutional neural network. Eur Radiol 30(2):1264–1273
Association CSoHCM, Association CSoGCM (2020) Chinese Society of Infectious Diseases CMA. Consensus on the diagnosis and treatment of hepatic fibrosis (2019). J Dig Dis 21(3): 127–138
Li Q, Yu B, Tian X, Cui X, Zhang R, Guo Q (2020) Deep residual nets model for staging liver fibrosis on plain CT images. Int J Comput Assist Radiol Surg 15(8):1399–1406
Kojiro M, Shimamatsu K, Kage M (1995) Pathomorphologic comparison of hepatitis C virus-related and hepatitis B virus-related cirrhosis bearing hepatocellular carcinoma. Princess Takamatsu Symp 25:179–184
Tampi C (2012) Pathology for the HPB Surgeon. Indian J Surg 74(1):67–72
Geng XX, Lin JM, Yang XX, Huang RG, Jiang N (2009) Comparison of liver pathohistological and clinical characteristics between chronic HBV carriers and chronic hepatitis B patients with mild elevation in ALT. Zhonghua Gan Zang Bing Za Zhi 17(10):735–739
He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. In: Conference proceedings: proceedings of the IEEE conference on computer vision and pattern recognition, pp 770–778
Mathew J, Pang CK, Luo M, Leong WH (2018) Classification of imbalanced data by oversampling in kernel space of support vector machines. IEEE Trans Neural Netw Learn Syst 29(9):4065–4076
Xie YN, Yu L, Guan GH, He YJ (2018) An overlapping cell image synthesis method for imbalance data. Anal Cell Pathol (Amst) 2018:7919503
Wang K, Lu X, Zhou H, Gao Y, Zheng J, Tong M, Wu C, Liu C, Huang L, Jiang T, Meng F, Lu Y, Ai H, Xie XY, Yin LP, Liang P, Tian J, Zheng R (2019) Deep learning Radiomics of shear wave elastography significantly improved diagnostic performance for assessing liver fibrosis in chronic hepatitis B: a prospective multicentre study. Gut 68(4):729–741
Warrens MJ (2013) Conditional inequalities between Cohen’s kappa and weighted kappas. Stat Methodol 10(1):14–22
Vilar-Gomez E, Chalasani N (2018) Non-invasive assessment of non-alcoholic fatty liver disease: clinical prediction rules and blood-based biomarkers. J Hepatol 68(2):305–315
Dai YN, Tu YX, Meng D, Chen MJ, Zhang JJ, Gong YH, Tong YX, Wang MS, Pan HY, Huang HJ (2019) Serum proteomic changes as candidate biomarkers of intermediate liver fibrosis in chronic hepatitis B infection. OMICS 23(3):167–179
Yen YH, Kuo FY, Kee KM, Chang KC, Tsai MC, Hu TH, Lu SN, Wang JH, Hung CH, Chen CH (2018) APRI and FIB-4 in the evaluation of liver fibrosis in chronic hepatitis C patients stratified by AST level. PLoS One 13(6): e0199760
Papadopoulos N, Vasileiadi S, Papavdi M, Sveroni E, Antonakaki P, Dellaporta E, Koutli E, Michalea S, Manolakopoulos S, Koskinas J, Deutsch M (2019) Liver fibrosis staging with combination of APRI and FIB-4 scoring systems in chronic hepatitis C as an alternative to transient elastography. Ann Gastroenterol 32(5):498–503
Chrostek L, Przekop D, Gruszewska E, Gudowska-Sawczuk M, Cylwik B (2019) Noninvasive indirect markers of liver fibrosis in alcoholics. Biomed Res Int 2019:3646975
Yang M, Jiang L, Wang Y, Li X, Zou Z, Han T, Nan Y, Lu F, Zhao J (2019) Step layered combination of noninvasive fibrosis models improves diagnostic accuracy of advanced fibrosis in nonalcoholic fatty liver disease. J Gastrointestin Liver Dis 28(3):289–296
Pickhardt PJ, Graffy PM, Said A, Jones D, Welsh B, Zea R, Lubner MG (2019) Multiparametric CT for noninvasive staging of hepatitis C virus-related liver fibrosis: correlation with the [2]histopathologic fibrosis score. AJR Am J Roentgenol 212(3):547–553
Lubner MG, Jones D, Said A, Kloke J, Lee S, Pickhardt PJ (2018) Accuracy of liver surface nodularity quantification on MDCT for staging hepatic fibrosis in patients with hepatitis C virus. Abdom Radiol (NY) 43(11):2980–2986
Pickhardt PJ, Malecki K, Hunt OF, Beaumont C, Kloke J, Ziemlewicz TJ, Lubner MG (2017) Hepatosplenic volumetric assessment at MDCT for staging liver fibrosis. Eur Radiol 27(7):3060–3068
Parola M, Pinzani M (2019) Liver fibrosis: pathophysiology, pathogenetic targets and clinical issues. Mol Aspects Med 65:37–55
Zhang CY, Yuan WG, He P, Lei JH, Wang CX (2016) Liver fibrosis and hepatic stellate cells: etiology, pathological hallmarks and therapeutic targets. World J Gastroenterol 22(48):10512–10522
Li H (2020) Angiogenesis in the progression from liver fibrosis to cirrhosis and hepatocelluar carcinoma. Expert Rev Gastroenterol Hepatol 15(3):217–233
Chiu NC, Su CW, Liu CA, Huang YH, Chiou YY (2017) Interval to vascularization development in cirrhotic precursor nodules in patients with hepatitis B and C virus co-infections. PLoS One 12(6): e0178841
Funding
Funding was provided by National Natural Science Foundation of China (Grant Nos. 81771893, 81471718).
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Li, Q., Kang, H., Zhang, R. et al. Non-invasive precise staging of liver fibrosis using deep residual network model based on plain CT images. Int J CARS 17, 627–637 (2022). https://doi.org/10.1007/s11548-022-02573-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11548-022-02573-8