Abstract
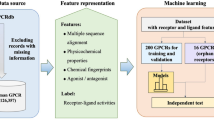
There are many new and potential drug targets in G protein-coupled receptors (GPCRs) without sufficient ligand associations, and accurately predicting and interpreting ligand bioactivities is vital for screening and optimizing hit compounds targeting these GPCRs. To efficiently address the lack of labeled training samples, we proposed a multi-task regression learning with incoherent sparse and low-rank patterns (MTR-ISLR) to model ligand bioactivities and identify their key substructures associated with these GPCRs targets. That is, MTR-ISLR intends to enhance the performance and interpretability of models under a small size of available training data by introducing homologous GPCR tasks. Meanwhile, the low-rank constraint term encourages to catch the underlying relationship among homologous GPCR tasks for greater model generalization, and the entry-wise sparse regularization term ensures to recognize essential discriminative substructures from each task for explanative modeling. We examined MTR-ISLR on a set of 31 important human GPCRs datasets from 9 subfamilies, each with less than 400 ligand associations. The results show that MTR-ISLR reaches better performance when compared with traditional single-task learning, deep multi-task learning and multi-task learning with joint feature learning-based models on most cases, where MTR-ISLR obtains an average improvement of 7% in correlation coefficient (r2) and 12% in root mean square error (RMSE) against the runner-up predictors. The MTR-ISLR web server appends freely all source codes and data for academic usages.1)
Similar content being viewed by others
Explore related subjects
Discover the latest articles and news from researchers in related subjects, suggested using machine learning.References
Sriram K, Insel P A. G protein-coupled receptors as targets for spproved drugs: how many targets and how many drugs? Molecular Pharmacology, 2018, 93(4): 251–258
Hauser A S, Attwood M M, Raskandersen M, Schioth H B, Gloriam D E. Trends in GPCR drug discovery: new agents, targets and indications. Nature Reviews Drug Discovery, 2017, 16(12): 829–842
Berman H M, Westbrook J, Feng Z, Gilliland G, Bhat T, Weissig H, Shindyalov I N, Bourne P E. The protein data bank. Nucleic Acids Research, 2010, 28: 235–242
Chan W K B, Zhang H, Yang J, Brender J R, Hur J, Ozgur A, Zhang Y. GLASS: a comprehensive database for experimentally validated GPCR-ligand associations. Bioinformatics, 2015, 31(18): 3035–3042
Blum L C, Reymond J. 970 million druglike small molecules for virtual screening in the chemical universe database GDB-13. Journal of the American Chemical Society, 2009, 131(25): 8732–8733
Wang C-C, Zhao Y, Chen X. Drug-pathway association prediction: from experimental results to computational models. Briefings in Bioinformatics, 2020
Lee D. CONET: a virtual human system-centered platform for drug discovery. Frontiers of Computer Science, 2018, 12(1): 1–3
Cherkasov A, Muratov E N, Fourches D, Varnek A, Baskin I I, Cronin M T D, Dearden J C, Gramatica P, Martin Y C, Todeschini R. QSAR modeling: Where have you been? Where are you going to? Journal of Medicinal Chemistry, 2014, 57(12): 4977–5010
Ceretomassague A, Ojeda M J, Valls C, Mulero M, Garciavallve S, Pujadas G. Molecular fingerprint similarity search in virtual screening. Methods, 2015, 71: 58–63
Melville J L, Burke E K, Hirst J D. Machine learning in virtual screening. Combinatorial Chemistry High Throughput Screening, 2009, 12(4): 332–343
Wallach I, Dzamba M, Heifets A. AtomNet: A deep convolutional neural network for bioactivity prediction in structure-based drug discovery. Mathematische Zeitschrift, 2015, 47(1): 34–46
Winkler D A, Le T C. Performance of deep and shallow neural networks, the universal approximation theorem, activity cliffs, and QSAR. Molecular Informatics, 2016, 36(1-2)
Lavecchia A. Deep learning in drug discovery: opportunities, challenges and future prospects. Drug Discovery Today, 2019, 24(10): 2017–2032
Ramsundar B, Liu B, Wu Z, Verras A, Tudor M, Sheridan R P, Pande V. Is multitask deep learning practical for pharma? Journal of Chemical Information and Modeling, 2017, 57(8): 2068–2076
Xu Y, Ma J, Liaw A, Sheridan R P, Svetnik V. Demystifying multitask deep neural networks for quantitative structure-activity relationships. Journal of Chemical Information and Modeling, 2017, 57(10): 2490–2504
Unterthiner T, Mayr A, Klambauer G, Steijaert M, Wegner J K, Ceulemans H, Hochreiter S. Deep learning as an opportunity in virtual screening. In: Proceedings of the Deep Learning Workshop at NIPS. 2014, 1–9
Ma J, Sheridan R, Liaw A, Dahl G, Svetnik V. Deep neural nets as a method for quantitative structure-activity relationships. Journal of Chemical Information and Modeling, 2015, 55(2): 263–274
Duvenaud D, Maclaurin D, Aguilera-Iparraguirre J, Gómez-Bombarelli R, Hirzel T, Aspuru-Guzik A, Adams R. Convolutional networks on graphs for learning molecular fingerprints. Advances in Neural Information Processing Systems (NIPS), 2015
Wu J, Zhang Q, Wu W, Pang T, Hu H, Chan W K B, Ke X, Zhang Y. WDL-RF: predicting bioactivities of ligand molecules acting with G protein-coupled receptors by combining weighted deep learning and random forest. Bioinformatics, 2018, 34: 2271–2282
Wu J, Liu B, Chan W K B, Wu W, Pang T, Hu H, Yan S, Ke X, Zhang Y. Precise modelling and interpretation of bioactivities of ligands targeting G protein-coupled receptors. Bioinformatics, 2019, 35: i324–i332
Dahl G E, Jaitly N, Salakhutdinov R. Multi-task neural networks for QSAR predictions. Computer Science, 2014
Chen L, Shao K, Long X, Wang L. Multi-task regression learning for survival analysis via prior information guided transductive matrix completion. Frontiers of Computer Science, 2020, 14(5): 97–110
Wu J, Sun Y, Chan W K B, Zhu Y, Zhu W, Huang W, Hu H, Yan S, Pang T, Ke X. Homologous G protein-coupled receptors boost the modeling and interpretation of bioactivities of ligand molecules. Journal of Chemical Information and Modeling, 2020, 60(3): 1865–1875
Simoes R S, Maltarollo V G, Oliveira P R, Honorio K M. Transfer and multi-task learning in QSAR modeling: advances and challenges. Frontiers in Pharmacology, 2018, 9: 74
Chen J, Liu J, Ye J. Learning incoherent sparse and low-rank patterns from multiple tasks. In: Proceedings of the 16th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining. 2010, 1179–1187
Bairoch A M, Apweiler R, Wu C H, Barker W C, Boeckmann B, Ferro S, Gasteiger E, Huang H, Lopez R, Magrane M. The universal protein resource (UniProt). Nucleic Acids Research, 2004, 33: 154–159
Wang Y, Xiao J, Suzek TO, Zhang J, Wang J, Bryant S H. PubChem: a public information system for analyzing bioactivities of small molecules. Nucleic Acids Research, 2009, 37: 623–633
Nesterov Y. Introductory lectures on convex optimization: a basic course. 1st ed. Boston: Springer Publishing Company, 2014
Zhou J, Chen J, Ye J. MALSAR: multi-task learning via structural regularization. Arizona State University, 2011, 21
Zhou J, Liu J, Narayan V A, Ye J. Modeling disease progression via fused sparse group lasso. In: Proceedings of the 18th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining. 2012, 1095–1103
Fang Y, Kenakin T P, Liu C. Editorial: orphan GPCRs as emerging drug targets. Frontiers in Pharmacology, 2015, 6: 295
Zhang L, Nothacker H-P, Bohn L, Civelli O. Pharmacological characterization of a selective agonist for Bombesin Receptor Subtype-3. Biochemical and Biophysical Research Communications, 2009, 387(2): 283–288
Acknowledgements
This work was supported in part by the National Natural Science Foundation of China (Grant Nos. 61872198, 61971216, 81771478, 81973512), the Basic Research Program of Science and Technology Department of Jiangsu Province (BK20201378), the Natural Science Foundation of the Higher Education Institutions of Jiangsu Province (18KJB416005), and the Natural Science Foundation of Nanjing University of Posts and Telecommunications (NY218092). We thank all the people who have contributed to the system in a variety of ways.
Author information
Authors and Affiliations
Corresponding author
Additional information
Jiansheng Wu received his PhD degree in biomedical engineering from Southeast University, China in 2009. Currently he is an associate professor at Nanjing University of Posts and Telecommunications (NJUPT), China. His research interests are mainly AI in drug discovery, bioinformatics and FPGA accelerator.
Chuangchuang Lan is now studying for his MS degree in biomedical engineering at Nanjing University of Posts and Telecommunications, China. His main research interests are machine learning and bioinformatics.
Xuelin Ye is now studying for his BS degree at the University of Warwick, UK. Her main research interest is machine learning.
Jiale Deng is now studying for his BS degree at Modern Economics & Management College, Jiangxi University of Finance and Economic, China. His main research interests are machine learning and human resource management.
Wanqing Huang received her MS degree from Nanjing University of Posts and Telecommunications, China. Her main research interest is machine learning.
Xueni Yang is now studying for her BS degree at Nanjing University of Posts and Telecommunications, China in biomedical engineering. Her main research interests are machine learning and bioinformatics.
Yanxiang Zhu is the chief technical officer of VeriMake Research, China. His main research interests are embedded system, human-computer interaction and FPGA accelerator.
Haifeng Hu received his PhD degree from Nanjing University of Posts and Telecommunications (NJUPT), China in 2007. Currently, he is a professor at NJUPT. His research interests include large-scale similarity search, wireless sensor networks, wireless networking and distributed systems.
1) http://noveldelta.com/MTR_ISLR
Supporting information
The supporting information is available online at journal.hep.com.cn and link.springer.com.
Electronic Supplementary Material
11704_2021_478_MOESM1_ESM.pdf
Disclosing incoherent sparse and low-rank patterns inside homologous GPCR tasks for better modelling of ligand bioactivities
11704_2021_478_MOESM3_ESM.pdf
Disclosing incoherent sparse and low-rank patterns inside homologous GPCR tasks for better modelling of ligand bioactivities
Rights and permissions
About this article
Cite this article
Wu, J., Lan, C., Ye, X. et al. Disclosing incoherent sparse and low-rank patterns inside homologous GPCR tasks for better modelling of ligand bioactivities. Front. Comput. Sci. 16, 164322 (2022). https://doi.org/10.1007/s11704-021-0478-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11704-021-0478-6