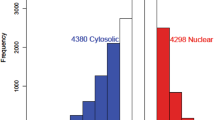
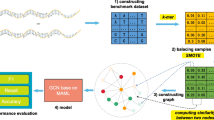
Abstract
The spatial distribution pattern of long non-coding RNA (lncRNA) in cell is tightly related to their function. With the increment of publicly available subcellular location data, a number of computational methods have been developed for the recognition of the subcellular localization of lncRNA. Unfortunately, these computational methods suffer from the low discriminative power of redundant features or overfitting of oversampling. To address those issues and enhance the prediction performance, we present a support vector machine-based approach by incorporating mutual information algorithm and incremental feature selection strategy. As a result, the new predictor could achieve the overall accuracy of 91.60%. The highly automated web-tool is available at lin-group.cn/server/iLoc-LncRNA(2.0)/website. It will help to get the knowledge of lncRNA subcellular localization.
Similar content being viewed by others
Explore related subjects
Discover the latest articles and news from researchers in related subjects, suggested using machine learning.References
Chiu H S, Somvanshi S, Patel E, Chen T W, Singh V P, Zorman B, Patil S L, Pan Y, Chatterjee S S, Cancer Genome Atlas Research N, Sood A K, Gunaratne P H, Sumazin P. Pan-cancer analysis of lncRNA regulation supports their targeting of cancer genes in each tumor context. Cell Reports, 2018, 23(1): 297–312.e12
Ji J, Tang J, Xia KJ, Jiang R. LncRNA in tumorigenesis microenvironment. Current Bioinformatics, 2019, 14(7): 640–641
Guo C J, Xu G, Chen L L. Mechanisms of long noncoding RNA nuclear retention. Trends in Biochemical Sciences, 2020, 45(11): 947–960
Chowdhury M R, Basak J, Bahadur R P. Elucidating the functional role of predicted miRNAs in post-transcriptional gene regulation along with symbiosis in medicago truncatula. Current Bioinformatics, 2020, 15(2): 108–120
Cheng L, Hu Y, Sun J, Zhou M, Jiang Q. DincRNA: a comprehensive web-based bioinformatics toolkit for exploring disease associations and ncRNA function. Bioinformatics, 2018, 34(11): 1953–1956
Cheng L, Wang P, Tian R, Wang S, Guo Q, Luo M, Zhou W, Liu G, Jiang H, Jiang Q. LncRNA2Target v2.0: a comprehensive database for target genes of lncRNAs in human and mouse. Nucleic Acids Research, 2019, 47(D1): D140–D144
Jiang Q, Ma R, Wang J, Wu X, Jin S, Peng J, Tan R, Zhang T, Li Y, Wang Y. LncRNA2Function: a comprehensive resource for functional investigation of human lncRNAs based on RNA-seq data. BMC Genomics, 2015, 16(3): 1–11
Jiang Q, Wang J, Wu X, Ma R, Zhang T, Jin S, Han Z, Tan R, Peng J, Liu G, Li Y, Wang Y. LncRNA2Target: a database for differentially expressed genes after lncRNA knockdown or overexpression. Nucleic Acids Research, 2015, 43(Database issue): D193–196
Jiang Q, Wang J, Wang Y, Ma R, Wu X, Li Y. TF2LncRNA: identifying common transcription factors for a list of lncRNA genes from ChIP-Seq data. Biomed Research International, 2014, 2014: 317642
Ning L, Cui T, Zheng B, Wang N, Luo J, Yang B, Du M, Cheng J, Dou Y, Wang D. MNDR v3.0: mammal ncRNA-disease repository with increased coverage and annotation. Nucleic Acids Research, 2021, 49(D1): D160–d164
Mora-Marquez F, Luis Vazquez-Poletti J, Chano V, Collada C, Soto A, Lopez de Heredia U. Hardware performance evaluation of de novo transcriptome assembly software in amazon elastic compute cloud. Current Bioinformatics, 2020, 15(5): 420–430
Hu B, Zheng L, Long C, Song M, Li T, Yang L, Zuo Y. EmExplorer: a database for exploring time activation of gene expression in mammalian embryos. Open Biology, 2019, 9(6): 190054
Zhu X, Li H D, Guo L, Wu F X, Wang J. Analysis of single-cell RNA-seq data by clustering approaches. Current Bioinformatics, 2019, 14(4): 314–322
Zhang T, Tan P, Wang L, Jin N, Li Y, Zhang L, Yang H, Hu Z, Zhang L, Hu C, Li C, Qian K, Zhang C, Huang Y, Li K, Lin H, Wang D. RNALocate: a resource for RNA subcellular localizations. Nucleic Acids Research, 2017, 45(D1): D135–D138
Mas-Ponte D, Carlevaro-Fita J, Palumbo E, Hermoso Pulido T, Guigo R, Johnson R. LncATLAS database for subcellular localization of long noncoding RNAs. RNA, 2017, 23(7): 1080–1087
Wen X, Gao L, Guo X, Li X, Huang X, Wang Y, Xu H, He R, Jia C, Liang F. lncSLdb: a resource for long non-coding RNA subcellular localization. Database (Oxford), 2018, 2018: 1–6
Gudenas B L, Wang L. Prediction of LncRNA subcellular localization with deep learning from sequence features. Science Reports, 2018, 8(1): 16385
Zhao T, Hu Y, Peng J, Cheng L. DeepLGP: a novel deep learning method for prioritizing lncRNA target genes. Bioinformatics, 2020, 36(16): 4466–4472
Zhao T, Hu Y, Cheng L. Deep-DRM: a computational method for identifying disease-related metabolites based on graph deep learning Approaches. Briefings in Bioinformatics, 2020, 22(4): bbaa212
Wu B, Zhang H, Lin L, Wang H, Gao Y, Zhao L, Chen Y-P P, Chen R, Gu L. A similarity searching system for biological phenotype images using deep convolutional encoder-decoder architecture. Current Bioinformatics, 2019, 14(7): 628–639
Charoenkwan P, Nantasenamat C, Hasan M M, Shoombuatong W. Meta-iPVP: a sequence-based meta-predictor for improving the prediction of phage virion proteins using effective feature representation. Journal of Computer-Aided Molecular Design, 2020, 34(10): 1105–1116
Liu K, Cao L, Du P, Chen W. im6A-TS-CNN: identifying the N(6)-methyladenine site in multiple tissues by using the convolutional neural network. Molecular Therapy-Nucleic Acids, 2020, 21: 1044–1049
Zuckerman B, Ulitsky I. Predictive models of subcellular localization of long RNAs. RNA, 2019, 25(5): 557–572
Dong Y M, Bi J H, He Q E, Song K. ESDA: an improved approach to accurately identify human snoRNAs for precision cancer therapy. Current Bioinformatics, 2020, 15(1): 34–40
Cao Z, Pan X, Yang Y, Huang Y, Shen H B. The lncLocator: a subcellular localization predictor for long non-coding RNAs based on a stacked ensemble classifier. Bioinformatics, 2018, 34(13): 2185–2194
Su Z D, Huang Y, Zhang Z Y, Zhao Y W, Wang D, Chen W, Chou K C, Lin H. iLoc-lncRNA: predict the subcellular location of lncRNAs by incorporating octamer composition into general PseKNC. Bioinformatics, 2018, 34(24): 4196–4204
Ahmad A, Lin H, Shatabda S. Locate-R: subcellular localization of long non-coding RNAs using nucleotide compositions. Genomics, 2020, 112(3): 2583–2589
Feng S, Liang Y, Du W, Lv W, Li Y. LncLocation: efficient subcellular location prediction of long non-coding RNA-based multi-source heterogeneous feature fusion. International Journal of Molecular Sciences, 2020, 21(19): 7271
Wang Y, Shi F, Cao L, Dey N, Wu Q, Ashour A S, Sherratt R S, Rajinikanth V, Wu L. Morphological segmentation analysis and texture-based support vector machines classification on mice liver fibrosis microscopic images. Current Bioinformatics, 2019, 14(4): 282–294
Pruitt K D, Tatusova T, Maglott D R. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Research, 2007, 35(Database issue): D61–65
Lai H Y, Zhang Z Y, Su Z D, Su W, Ding H, Chen W, Lin H. iProEP: a computational predictor for predicting promoter. Molecular Therapy-Nucleic Acids, 2019, 17: 337–346
Liu K, Chen W. iMRM: a platform for simultaneously identifying multiple kinds of RNA modifications. Bioinformatics, 2020, 36(11): 3336–3342
Hasan M M, Basith S, Khatun M S, Lee G, Manavalan B, Kurata H. Meta-i6mA: an interspecies predictor for identifying DNA N6-methyladenine sites of plant genomes by exploiting informative features in an integrative machine-learning framework. Briefings in Bioinformatics, 2020, 22(3): bbaa202
Manavalan B, Basith S, Shin T H, Wei L, Lee G. Meta-4mCpred: a sequence-based meta-predictor for accurate DNA 4mC site prediction using effective feature representation. Molecular Therapy-Nucleic Acids, 2019, 16: 733–744
Basith S, Manavalan B, Shin T H, Lee G. SDM6A: a web-based integrative machine-learning framework for predicting 6mA sites in the rice genome. Molecular Therapy-Nucleic Acids, 2019, 18: 131–141
Zheng L, Huang S, Mu N, Zhang H, Zhang J, Chang Y, Yang L, Zuo Y. RAACBook: a web server of reduced amino acid alphabet for sequence-dependent inference by using Chou’s five-step rule. Database (Oxford), 2019
Zhang Z Y, Yang Y H, Ding H, Wang D, Chen W, Lin H. Design powerful predictor for mRNA subcellular location prediction in Homo sapiens. Briefings in Bioinformatics, 2021, 22(1): 526–535
Zhang J, Liu B. A review on the recent developments of sequence-based protein feature extraction methods. Current Bioinformatics, 2019, 14(3): 190–199
Liang P F, Yang W R, Chen X, Long C S, Zheng L, Li H S, Zuo Y C. Machine learning of single-cell transcriptome highly identifies mRNA signature by comparing F-score selection with DGE analysis. Molecular Therapy-Nucleic Acids, 2020, 20: 155–163
Liu K, Chen W, Lin H. XG-PseU: an eXtreme Gradient Boosting based method for identifying pseudouridine sites. Molecular Genetics and Genomics, 2020, 295(1): 13–21
Guo X, Gao L, Wang Y, Chiu D K Y, Wang B, Deng Y, Wen X. Large-scale investigation of long noncoding RNA secondary structures in human and mouse. Current Bioinformatics, 2018, 13(5): 450–460
Zhang D, Xu Z C, Su W, Yang Y H, Lv H, Yang H, Lin H. iCarPS: a computational tool for identifying protein carbonylation sites by novel encoded features. Bioinformatics, 2021, 37(2): 171–177
Wang S P, Zhang Q, Lu J, Cai Y D. Analysis and prediction of nitrated tyrosine sites with the mRMR method and support vector machine algorithm. Current Bioinformatics, 2018, 13(1): 3–13
Peng H, Long F, Ding C. Feature selection based on mutual information: criteria of max-dependency, max-relevance, and min-redundancy. IEEE Transactions on Pattern Analysis and Machine Intelligence, 2005, 27(8): 1226–1238
Chen J, Zhao J, Yang S, Chen Z, Zhang Z. Prediction of protein ubiquitination sites in arabidopsis thaliana. Current Bioinformatics, 2019, 14(7): 614–620
Charoenkwan P, Nantasenamat C, Hasan M M, Shoombuatong W. iTTCA-Hybrid: improved and robust identification of tumor T cell antigens by utilizing hybrid feature representation. Analytical Biochemistry, 2020, 599: 113747
Jiang Q, Wang G, Jin S, Li Y, Wang Y. Predicting human microRNA-disease associations based on support vector machine. International Journal of Dato Mining and Bioinformatics, 2013, 8(3): 282–293
Chang C C, Lin C J. LIBSVM: a library for support vector machines. ACM Transactions on Intelligent Systems and Technology, 2011, 2(3): 27
Wei L, He W, Malik A, Su R, Cui L, Manavalan B. Computational prediction and interpretation of cell-specific replication origin sites from multiple eukaryotes by exploiting stacking framework. Briefings in Bioinformatics, 2021, 22(4): bbaa275
Hasan M M, Manavalan B, Shoombuatong W, Khatun M S, Kurata H. i4mC-Mouse: improved identification of DNA N4-methylcytosine sites in the mouse genome using multiple encoding schemes. Computational and Structural Biotechnology Journal, 2020, 18: 906–912
Charoenkwan P, Yana J, Schaduangrat N, Nantasenamat C, Hasan M M, Shoombuatong W. iBitter-SCM: identification and characterization of bitter peptides using a scoring card method with propensity scores of dipeptides. Genomics, 2020, 112(4): 2813–2822
Charoenkwan P, Chiangjong W, Lee V S, Nantasenamat C, Hasan M M, Shoombuatong W. Improved prediction and characterization of anticancer activities of peptides using a novel flexible scoring card method. Scientific Reports, 2021, 11(1): 1–13
Charoenkwan P, Kanthawong S, Nantasenamat C, Hasan M M, Shoombuatong W. iDPPIV-SCM: a sequence-based predictor for identifying and analyzing dipeptidyl peptidase IV (DPP-IV) inhibitory peptides using a scoring card method. Journal of Proteome Research, 2020, 19(10): 4125–4136
Charoenkwan P, Kanthawong S, Nantasenamat C, Hasan M M, Shoombuatong W. iAMY-SCM: improved prediction and analysis of amyloid proteins using a scoring card method with propensity scores of dipeptides. Genomics, 2021, 113(1): 689–698
Charoenkwan P, Kanthawong S, Schaduangrat N, Yana J, Shoombuatong W. PVPred-SCM: improved prediction and analysis of phage virion proteins using a scoring card method. Cells, 2020, 9(2): 353
Charoenkwan P, Nantasenamat C, Hasan M M, Shoombuatong W. iTTCA-Hybrid: improved and robust identification of tumor T cell antigens by utilizing hybrid feature representation. Analytical Biochemistry, 2020, 599: 113747
Charoenkwan P, Shoombuatong W, Lee H C, Chaijaruwanich J, Huang H L, Ho S Y. SCMCRYS: predicting protein crystallization using an ensemble scoring card method with estimating propensity scores of P-collocated amino acid pairs. PLoS ONE, 2013, 8(9): e72368
Charoenkwan P, Yana J, Nantasenamat C, Hasan M M, Shoombuatong W. iUmami-SCM: a novel sequence-based predictor for prediction and analysis of umami peptides using a scoring card method with propensity scores of dipeptides. Journal of Chemical Information and Modeling, 2020, 60(12): 6666–6678
Long H, Sun Z, Li M, Fu H Y, Lin M C. Predicting protein phosphorylation sites based on deep learning. Current Bioinformatics, 2020, 15(4): 300–308
Cheng L. Computational and biological methods for gene therapy. Current Gene Therapy, 2019, 19(4): 210–210
Cheng L, Hu Y. Human disease system biology. Current Gene Therapy, 2018, 18(5): 255–256
Kuang L, Zhao H, Wang L, Xuan Z, Pei T. A novel approach based on point cut set to predict associations of diseases and LncRNAs. Current Bioinformatics, 2019, 14(4): 333–343
Chen W, Feng P, Song X, Lv H, Lin H. iRNA-m7G: identifying N(7)-methylguanosine sites by fusing multiple features. Molecular Therapy Nucleic Acids, 2019, 18: 269–274
Liu D, Li G, Zuo Y. Function determinants of TET proteins: the arrangements of sequence motifs with specific codes. Briefings in Bioinformatics, 2019, 20(5): 1826–1835
Zheng L, Liu D, Yang W, Yang L, Zuo Y. RaacLogo: a new sequence logo generator by using reduced amino acid clusters. Briefings in Bioinformatics, 2021, 22(3): bbaa096
Bailey T L. DREME: motif discovery in transcription factor ChIP-seq data. Bioinformatics, 2011, 27(12): 1653–1659
Ginestet C. ggplot2: elegant graphics for data analysis. Journal of the Royal Statistical Society Series a-Statistics in Society, 2011, 174: 245–245
Acknowledgements
This work was supported by the National Nature Scientific Foundation of China (Grant No. 61772119), Sichuan Provincial Science Fund for Distinguished Young Scholars (2020JDJQ0012).
Author information
Authors and Affiliations
Corresponding author
Additional information
Zhao-Yue Zhang received the MS degree in Biophysics from University of Electronic Science and Technology of China, China. She is a research assistant of Center for Informational Biology and the Key Laboratory for NeuroInformation of Ministry of Education in University of Electronic Science and Technology of China. Her research interests are bioinformatics, machine learning and RNA subcellular localization.
Zi-Jie Sun is a graduate student at the Center for Informational Biology, University of Electronic Science and Technology of China, China. Her research interests are bioinformatics, statistical analysis and drug repositioning.
Yu-He Yang is a graduate student at the Center for Informational Biology, University of Electronic Science and Technology of China, China. Her research interests are bioinformatics, machine learning and RNA methylation.
Hao Lin is a professor at the Center for Informational Biology, University of Electronic Science and Technology of China, China. His research is in the areas of bioinformatics and system biology.
Electronic Supplementary Material
Rights and permissions
About this article
Cite this article
Zhang, ZY., Sun, ZJ., Yang, YH. et al. Towards a better prediction of subcellular location of long non-coding RNA. Front. Comput. Sci. 16, 165903 (2022). https://doi.org/10.1007/s11704-021-1015-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11704-021-1015-3