Abstract
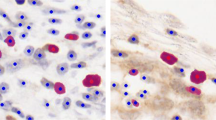
This paper presents segmentation of p53immunostained tissue images of oral squamous cell carcinoma that consist of cell nuclei segmentation and splitting of overlapping/touching cell structures. In segmentation, the entropy thresholding has been adopted in which the optimum threshold value to each color component of the image is obtained by maximizing the global entropy computed from its gray-level co-occurrence matrix. The segmented image consists of isolated cells and complex nuclei structures. A novel complex nuclei structure detection algorithm is proposed to identify overlapped nuclei structures, which have been further resolved by watershed transform. The performance of the segmentation technique is evaluated using the quantitative metrics, namely mean absolute difference (MAD), dice coefficient (DC) and accuracy. Global entropy thresholding-based segmentation technique achieved the best MAD of 0.478, DC of 0.967 and accuracy of 0.970 compared to state-of-art techniques such as otsu and active contour. Extensive experimental results show that proposed complex nuclei structure detection-based overlapping/touching cells splitting algorithm effectively delineated nuclei with over- and under-segmentation rate of 0.49 %. Therefore, tissue image segmentation method presented has high potential in immunohistochemical (IHC) quantification and also can be easily generalized for images stained with other biomarkers.






Similar content being viewed by others
References
Chang, S.W., Abdul-Kareem, S., Merican, A.F., Zain, R.B.: Oral cancer prognosis based on clinicopathologic and genomic markers using a hybrid of feature selection and machine learning methods. BMC Bioinform. 14(170), 1–15 (2013)
Polanska, H., Raudenska, M., Gumulec, J., Sztalmachova, M., Adam, V., Kizek, R., Masarik, M.: Clinical significance of head and neck squamous cell cancer biomarkers. Oral Oncol. 50(3), 168–177 (2014)
Swaminathan, U., Joshua, E., Rao, U.K., Ranganathan, K.: Expression of p53 and cyclin D1 in oral squamous cell carcinoma and normal mucosa: an immunohistochemical study. J. Oral Maxillofac. Pathol (JOMFP) 16(2), 172 (2012)
Ebrahimi, M., Boldrup, L., Coates, P.J., Wahlin, Y.B., Bourdon, J.C., Nylander, K.: Expression of novel p53 isoforms in oral lichen planus. Oral Oncol. 44(2), 156–161 (2008)
Reibel, J.: Prognosis of oral pre-malignant lesions: significance of clinical, histopathological, and molecular biological characteristics. Crit. Rev. Oral Biol. Med. 14(1), 47–62 (2003)
Di Cataldo, S., Ficarra, E., Macii, E.: Computer-aided techniques for chromogenic immunohistochemistry: status and directions. Comput. Biol. Med. 42(10), 1012–1025 (2012)
Fuchs, T.J., Buhmann, J.M.: Computational pathology: challenges and promises for tissue analysis. Comput. Med. Imaging Graph. 35(7), 515–530 (2011)
Samsi, S., Krishnamurthy, A.K., Gurcan, M.N.: An efficient computational framework for the analysis of whole slide images: application to follicular lymphoma immunohistochemistry. J. Comput. Sci. 3(5), 269–279 (2012)
Wienert, S., Heim, D., Kotani, M., Lindequist, B., Stenzinger, A., Ishii, M., Hufnagl, P., Beil, M., Dietel, M., Denkert, C., et al.: Cognitionmaster: an object-based image analysis framework. Diagn. Pathol. 8(1), 34–34 (2013)
He, L., Long, L.R., Antani, S., Thoma, G.R.: Histology image analysis for carcinoma detection and grading. Comput. Methods Programs Biomed. 107(3), 538–556 (2012)
Masmoudi, H., Hewitt, S.M., Petrick, N., Myers, K.J., Gavrielides, M.A.: Automated quantitative assessment of HER-2/neu immunohistochemical expression in breast cancer. IEEE Trans. Med. Imaging 28(6), 916–925 (2009)
Di Cataldo, S., Ficarra, E., Acquaviva, A., Macii, E.: Automated segmentation of tissue images for computerized IHC analysis. Comput. Methods Programs Biomed. 100(1), 1–15 (2010)
Chang, C.I., Chen, K., Wang, J., Althouse, M.L.: A relative entropy-based approach to image thresholding. Pattern Recognit. 27(9), 1275–1289 (1994)
Chang, C.I., Du, Y., Wang, J., Guo, S.M., Thouin, P.: Survey and comparative analysis of entropy and relative entropy thresholding techniques. In: IEE Proceedings of the Vision, Image and Signal Processing, vol 153, pp. 837–850. IET (2006)
Sertel, O., Lozanski, G., Shana’ah, A., Gurcan, M.N.: Computer-aided detection of centroblasts for follicular lymphoma grading using adaptive likelihood-based cell segmentation. IEEE Trans. Biomed. Eng. 57(10), 2613–2616 (2010)
Mudrova, M., Prochazka, A.: Principal component analysis in image processing. In: Proceedings of the MATLAB Technical Computing Conference, Prague (2005)
Yimit, A., Hagihara, Y., Miyoshi, T., Hagihara, Y., Yimit, Q.: Fast method for two-dimensional renyi\(^{\prime }\)s entropy-based thresholding. Int. J. Comput. Sci. Eng. 4(2), 176–183 (2012)
Cloppet, F., Boucher, A.: Segmentation of complex nucleus configurations in biological images. Pattern Recognit. Lett. 31(8), 755–761 (2010)
Mouelhi, A., Sayadi, M., Fnaiech, F., Mrad, K., Romdhane, K.B.: Automatic image segmentation of nuclear stained breast tissue sections using color active contour model and an improved watershed method. Biomed. Signal Process. Control 8(5), 421–436 (2013)
Qi, X., Xing, F., Foran, D.J., Yang, L.: Robust segmentation of overlapping cells in histopathology specimens using parallel seed detection and repulsive level set. IEEE Trans. Biomed. Eng. 59(3), 754–765 (2012)
Yang, X., Li, H., Zhou, X.: Nuclei segmentation using marker-controlled watershed, tracking using mean-shift, and Kalman filter in time-lapse microscopy. IEEE Trans. Circuits Syst. I Regul. Pap. 53(11), 2405–2414 (2006)
Veta, M., van Diest, P.J., Kornegoor, R., Huisman, A., Viergever, M.A., Pluim, J.P.: Automatic nuclei segmentation in H&E stained breast cancer histopathology images. PLoS One 8(7), e70221 (2013)
Lu, C., Mahmood, M., Jha, N., Mandal, M.: Detection of melanocytes in skin histopathological images using radial line scanning. Pattern Recognit. 46(2), 509–518 (2013)
Zhang, L., Kong, H., Chin, C.T., Liu, S., Chen, Z., Wang, T., Chen, S.: Segmentation of cytoplasm and nuclei of abnormal cells in cervical cytology using global and local graph cuts. Comput. Med. Imaging Graph. 38(5), 369–380 (2014)
Di Cataldo, S., Ficarra, E., Acquaviva, A., Macii, E.: Achieving the way for automated segmentation of nuclei in cancer tissue images through morphology-based approach: a quantitative evaluation. Comput. Med. Imaging Graph. 34(6), 453–461 (2010)
Acknowledgments
The authors are expressing sincere thanks to Dr. Siow-Wee Chang, Faculty of Science, Institute of Biological Sciences, University of Malaya, for providing IHC-stained tissue images of OSCC used in this study. Special thanks to Dr. Anil Malleshi Betigeri, M.D., and Dr. Mathusudanan, M.D., Department of Pathology, Meenakashi Mission Hospital and Research Center, Madurai, Tamilnadu, India, for their supports in understanding the concepts and also for their valuable suggestions during this research work.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Shahul Hameed, K.A., Banumathi, A. & Ulaganathan, G. P53immunostained cell nuclei segmentation in tissue images of oral squamous cell carcinoma. SIViP 11, 363–370 (2017). https://doi.org/10.1007/s11760-016-0953-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11760-016-0953-y