Abstract
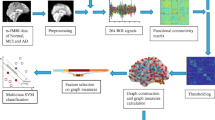
Functional magnetic resonance imaging (fMRI) is an imaging tool that is used to analyze the brain’s functions. Brain functional connectivity analysis based on fMRI signals often calculated correlations among time series in different areas of the brain. For FC analysis most prior research works generate the brain graphs based on linear correlations, however, the nonlinear behavior of the brain can lower the accuracy of such graphs. Usually, the Pearson correlation coefficient is used which has limitations in revealing nonlinear relationships. One of the proper methods for nonlinear analysis is the Kernel trick. This method maps the data into a high dimensional space and calculates the linear relations in a new space that is equivalent to the nonlinear relation in primary space. Also, it does not need to know the nonlinear dependency in the initial space. In this study, after constructing weighted undirected graphs of fMRI data based on AAL atlas, different kernels have been applied to calculate the kernelized correlation in normal and Alzheimer’s subjects. The determination of parameters has been done by two statistical methods. To compare the performance of Kernel correlation analysis, the global features of graphs are computed. Also, the non-parametric permutation test shows that kernelized correlation demonstrates a more significant statistical difference between groups in comparison to the simple linear correlation. In different kernel analysis, the best performance was for the third-degree polynomial kernel. The features strength, characteristic path length, local efficiency, transitivity, modularity, and small-worldness were significantly different for P value 0.01. Besides, comparison to random graphs and further analysis in the Occipital lobe confirmed the results.




Similar content being viewed by others
References
Alzheimer’s, A.: Alzheimer’s disease facts and figures. Alzheimer’s Dementia J. Alzheimer’s Assoc. 11(3), 332 (2015)
Samieri, C.: Epidemiology and risk factors of Alzheimer’s disease: a focus on diet. In: Perneczky, R.G. (ed.) Biomarkers for Preclinical Alzheimer’s Disease, vol. 137, pp. 15–42. Springer (2018)
Ashby, F.G.: An introduction to fMRI. In: Forstmann, B.U., Wagenmakers, E.-J. (eds.) An Introduction to Model-Based Cognitive Neuroscience, pp. 91–112. Springer, New York (2015)
Lee, M.H., Smyser, C.D., Shimony, J.S.: Resting-state fMRI: a review of methods and clinical applications. Am. J. Neuroradiol. 34(10), 1866 (2013)
Dai, Z., et al.: Identifying and mapping connectivity patterns of brain network hubs in Alzheimer’s disease. Cereb. Cortex 25(10), 3723–3742 (2014)
Zhan, Y., et al.: Longitudinal study of impaired intra-and inter-network brain connectivity in subjects at high risk for Alzheimer’s disease. J. Alzheimers Dis. 52(3), 913–927 (2016)
Greicius, M.D., Srivastava, G., Reiss, A.L., Menon, V.: Default-mode network activity distinguishes Alzheimer’s disease from healthy aging: evidence from functional MRI. Proc. Natl. Acad. Sci. 101(13), 4637–4642 (2004)
He, Y., et al.: Regional coherence changes in the early stages of Alzheimer’s disease: a combined structural and resting-state functional MRI study. Neuroimage 35(2), 488–500 (2007). https://doi.org/10.1016/j.neuroimage.2006.11.042
Zhang, H.-Y., et al.: Detection of PCC functional connectivity characteristics in resting-state fMRI in mild Alzheimer’s disease. Behav. Brain Res. 197(1), 103–108 (2009)
Cao, J., Worsley, K.: The geometry of correlation fields with an application to functional connectivity of the brain. Ann. Appl. Probab. 9(4), 1021–1057 (1999)
Vanderwal, T., Eilbott, J., Finn, E.S., Craddock, R.C., Turnbull, A., Castellanos, F.X.: Individual differences in functional connectivity during naturalistic viewing conditions,”. NeuroImage 157, 521–530 (2017)
Yoo, K., et al.: Connectome-based predictive modeling of attention: comparing different functional connectivity features and prediction methods across datasets. NeuroImage 167, 11–22 (2018)
Ochab, J.K., Tarnowski, W., Nowak, M.A., Chialvo, D.R.: On the pros and cons of using temporal derivatives to assess brain functional connectivity. NeuroImage 184, 577–585 (2019)
Benesty, J., Chen, J., Huang, Y., Cohen, I.: Pearson correlation coefficient. In: Noise Reduction in Speech Processing, Springer Topics in Signal Processing, vol. 2. Springer, Berlin, Heidelberg (2009). https://doi.org/10.1007/978-3-642-00296-0_5
Anzellotti, S., Fedorenko, E., Kell, A.J., Caramazza, A., Saxe, A.: Measuring and modeling nonlinear interactions between brain regions with fMRI bioRxiv, Article 074856 (2017)
Mesejo, P., Saillet, S., David, O., Bénar, C.G., Warnking, J.M., Forbes, F.: A differential evolution-based approach for fitting a nonlinear biophysical model to fMRI BOLD data. IEEE J. Select. Top. Signal Process. 10(2), 416–427 (2016)
Gultepe, E., He, B.: A linear/nonlinear characterization of resting state brain networks in FMRI time series. Brain Topogr. 26(1), 39–49 (2013)
Lindquist, M.A., Loh, J.M., Atlas, L.Y., Wager, T.D.: Modeling the hemodynamic response function in fMRI: efficiency, bias and mis-modeling. Neuroimage 45(1), S187–S198 (2009)
Marinazzo, D., Pellicoro, M., Stramaglia, S.: Kernel method for nonlinear Granger causality. Phys. Rev. Lett. 100(14), 144103 (2008)
Meszlényi, R.J., Hermann, P., Buza, K., Gál, V., Vidnyánszky, Z.: Resting state fMRI functional connectivity analysis using dynamic time warping. Front. Neurosci. 11, 75 (2017)
Bi, X.-A., Sun, Q., Zhao, J., Xu, Q., Wang, L.: nonlinear ICA analysis of resting-state fMRI in mild cognitive impairment. Front. Neurosci. 12, 413 (2018)
Hughes, J.A., Daley, M.: Finding nonlinear relationships in fmri time series with symbolic regression. In: Proceedings of the 2016 on Genetic and Evolutionary Computation Conference Companion, pp. 101–102 (2016)
Karanikolas, G., Giannakis, G.B., Slavakis, K., Leahy, R.M.: Multi-kernel based nonlinear models for connectivity identification of brain networks. In: IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), pp. 6315–6319. IEEE (2016)
Roth, V., Steinhage, V.: Nonlinear discriminant analysis using kernel functions. In: Advances in Neural Information Processing Systems, pp. 568–574 (2000)
Hardoon, D.R., Mourao-Miranda, J., Brammer, M., Shawe-Taylor, J.: Unsupervised analysis of fMRI data using kernel canonical correlation. NeuroImage 37(4), 1250–1259 (2007)
Sporns, O.: Graph theory methods: applications in brain networks. Dialogues Clin. Neurosci. 20(2), 111 (2018)
de Vos, F., et al.: A comprehensive analysis of resting state fMRI measures to classify individual patients with Alzheimer’s disease. Neuroimage 167, 62–72 (2018)
Rubinov, M., Sporns, O.: Complex network measures of brain connectivity: uses and interpretations. Neuroimage 52(3), 1059–1069 (2010)
Kepner, J., Gilbert, J.: Graph Algorithms in the Language of Linear Algebra. Society for Industrial & Applied Mathematics (2011)
Humphries, M.D., Gurney, K.: Network ‘small-world-ness’: a quantitative method for determining canonical network equivalence. PLoS ONE 3(4), e0002051 (2008)
Alam, M.A., Calhoun, V.D., Wang, Y.-P.: Identifying outliers using multiple kernel canonical correlation analysis with application to imaging genetics. Comput. Stat. Data Anal. 125, 70–85 (2018)
Hofmann, T., Schölkopf, B., Smola, A.J.: Kernel methods in machine learning. Ann. Stat. 36, 1171–1220 (2008)
Hofmann, M.: Support vector machines-kernels and the kernel trick. Notes 26, 3 (2006)
Kung, S.Y.: Kernel Methods and Machine Learning. Cambridge University Press, Cambridge (2014)
Towsley, A., Pakianathan, J., Douglass, D.H.: Correlation angles and inner products: application to a problem from physics. ISRN Appl. Math. 2011, 323864 (2011)
Scholkopf, B., Smola, A.J.: Learning with Kernels: Support Vector Machines, Regularization, Optimization, and Beyond. MIT Press, Cambridge (2001)
Vert, J.-P., Tsuda, K., Schölkopf, B.: A primer on kernel methods. Kernel Methods Comput. Biol. 47, 35–70 (2004)
Liu, Z., Zuo, M.J., Zhao, X., Xu, H.: An analytical approach to fast parameter selection of Gaussian RBF kernel for support vector machine. J. Inf. Sci. Eng. 31(2), 691–710 (2015)
Bilski, P.: Automated selection of kernel parameters in diagnostics of analog systems. Przegląd Elektrotechniczny 87(5), 9–13 (2011)
Garreau, D., Jitkrittum, W., Kanagawa, M.: Large sample analysis of the median heuristic. arXiv:1707.07269 (2017)
Gretton, A., et al.: Optimal kernel choice for large-scale two-sample tests. In: Advances in Neural Information Processing Systems, pp. 1205–1213 (2012)
Amami, R., Ayed, D.B., Ellouze, N.: Practical selection of SVM supervised parameters with different feature representations for vowel recognition. arXiv:1507.06020 (2015)
Lin, H.-T., Lin, C.-J.: A study on sigmoid kernels for SVM and the training of non-PSD kernels by SMO-type methods. Neural Comput. 3, 1–32 (2003)
Hua, X., et al.: MRI-based brain atrophy rates in ADNI phase 2: acceleration and enrichment considerations for clinical trials. Neurobiol. Aging 37, 26–37 (2016)
Yan, C., Zang, Y.: DPARSF: a MATLAB toolbox for “pipeline” data analysis of resting-state fMRI. Front. Syst. Neurosci. 4, 13 (2010)
Tzourio-Mazoyer, N., et al.: Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. NeuroImage 15(1), 273–289 (2002)
Mijalkov, M., Kakaei, E., Pereira, J.B., Westman, E., Volpe, G., Initiative, A.S.D.N.: BRAPH: a graph theory software for the analysis of brain connectivity. PloS ONE 12(8), e0178798 (2017)
Hilgetag, C.C., Goulas, A.: Is the brain really a small-world network? Brain Struct. Funct. 221(4), 2361–2366 (2016)
Coninck, J.C., Ferrari, F.A., Reis, A.S., Iarosz, K.C., Batista A.M., Viana, R.L.: Network properties of healthy and Alzheimer’s brains. arXiv:1905.11249 (2019)
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Ahmadi, H., Fatemizadeh, E. & Motie-Nasrabadi, A. fMRI functional connectivity analysis via kernel graph in Alzheimer’s disease. SIViP 15, 715–723 (2021). https://doi.org/10.1007/s11760-020-01789-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11760-020-01789-y