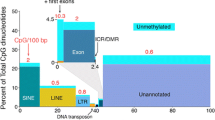
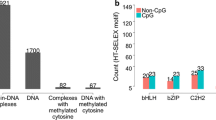
Abstract
Total DNA methylation rates are well known to vary widely between different metazoans. The phylogenetic distribution of this variation, however, has not been investigated systematically. We combine here publicly available data on methylcytosine content with the analysis of nucleotide compositions of genomes and transcriptomes of 78 metazoan species to trace the evolution of abundance and distribution of DNA methylation. The depletion of CpG and the associated enrichment of TpG and CpA dinucleotides are used to infer the intensity and localization of germline CpG methylation and to estimate its evolutionary dynamics. We observe a positive correlation of the relative methylation of CpG motifs with genome size. We tested this trend successfully by measuring total DNA methylation with LC/MS in orthopteran insects with very different genome sizes: house crickets, migratory locusts and meadow grasshoppers. We hypothesize that the observed correlation between methylation rate and genome size is due to a dependence of both variables from long-term effective population size and is driven by the accumulation of repetitive sequences that are typically methylated during periods of small population sizes. This process may result in generally methylated, large genomes such as those of jawed vertebrates. In this case, the emergence of a novel demethylation pathway and of novel reader proteins for methylcytosine may have enabled the usage of cytosine methylation for promoter-based gene regulation. On the other hand, persistently large populations may lead to a compression of the genome and to the loss of the DNA methylation machinery, as observed, e.g., in nematodes.







Similar content being viewed by others
Notes
References
Arndt PF, Burge CB, Hwa T (2003a) DNA sequence evolution with neighbor-dependent mutation. J Comput Biol 10:313–322
Arndt PF, Hwa T (2005) Identification and measurement of neighbor-dependent nucleotide substitution processes. Bioinformatics 21:2322–2328
Arndt PF, Petrov DA, Hwa T (2003b) Distinct changes of genomic biases in nucleotide substitution at the time of mammalian radiation. Mol Biol Evol 20:1887–1896
Bhutani N, Brady JJ, Damian M, Sacco A, Corbel SY, Blau HM (2010) Reprogramming towards pluripotency requires AID-dependent DNA demethylation. Nature 463:1042–1047
Bird AP (1980) DNA methylation and the frequency of CpG in animal DNA. Nucleic Acids Res 8:1499–1504
Bird AP, Taggart MH, Smith BA (1979) Methylated and unmethylated DNA compartments in the sea urchin genome. Cell 17:889–901
Boerjan B, Sas F, Ernst UR, Tobback J, Lemière F, Vandegehuchte MB, Janssen CR, Badisco L, Marchal E, Verlinden H, Schoofs L, Loof AD (2011) Locust phase polyphenism: Does epigenetic precede endocrine regulation? Gen Comp Endocrinol 173:120–128
Bogdanovic O, Veenstra GJC (2009) DNA methylation and methyl-CpG binding proteins: developmental requirements and function. Chromosoma 118:549–565
Charlesworth B (2009) Fundamental concepts in genetics: effective population size and patterns of molecular evolution and variation. Nat Rev Genet 10:195–205
Chen WJ, Ortí G, Meyer A (2004) Novel evolutionary relationship among four fish model systems. Trends Genet 20:424–431
Choi JK, Bae JB, Lyu J, Kim TY, Kim YJ (2009) Nucleosome deposition and DNA methylation at coding region boundaries. Genome Biol 10:R89
Choy JS, Wei S, Lee JY, Tan S, Chu S, Lee TH (2010) DNA methylation increases nucleosome compaction and rigidity. J Am Chem Soc 132:1782–1783
Clark SJ, Harrison J, Frommer M (1995) CpNpG methylation in mammalian cells. Nat Genetics 10:20–27
Consortium DG (2007) Evolution of genes and genomes on the Drosophila phylogeny. Nature 450:203–218
Conticello S (2008) The AID/APOBEC family of nucleic acid mutators. Genome Biol 9:229
Conticello SG, Thomas CJ, Petersen-Mahrt SK, Neuberger MS (2005) Evolution of the AID/APOBEC family of polynucleotide (deoxy)cytidine deaminases. Mol Biol Evol 22:367–377
Duret L, Galtier N (2000) The covariation between TpA deficiency, CpG deficiency, and G+C content of human isochores is due to a mathematical artifact. Mol Biol Evol 17:1620–1625
Elango N, Hunt BG, Goodisman MA, Yi SV (2009) DNA methylation is widespread and associated with differential gene expression in castes of the honeybee, Apis mellifera. Proc Natl Acad Sci USA 106:11206–11211
Elango N, Kim SH, Vigoda E, Yi SV (2008) Mutations of different molecular origins exhibit contrasting patterns of regional substitution rate variation. PLoS Comput Biol 4:e1000015
Elango N, Yi SV (2008) DNA methylation and structural and functional bimodality of vertebrate promoters. Mol Biol Evol 25:1602–1608
Eyre-Walker A, Keightley PD, Smith NGC, Gaffney D (2002) Quantifying the slightly deleterious mutation model of molecular evolution. Mol Biol Evol 19:2142–2149
Feng S, Cokus SJ, Zhang X, Chen PY, Bostick M, Goll MG, Hetzel J, Jain J, Strauss SH, Halpern ME, Ukomadu C, Sadler KC, Pradhan S, Pellegrini M, Jacobsen SE (2010) Conservation and divergence of methylation patterning in plants and animals. Proc Natl Acad Sci USA 107:8689–8694
Feschotte C, Keswani U, Ranganathan N, Guibotsy ML, Levine D (2009) Exploring repetitive DNA landscapes using REPCLASS, a tool that automates the classification of transposable elements in eukaryotic genomes. Genome Biol Evol 1:205–220
Feuerbach L, Lyngsø RB, Lengauer T, Hein J (2011) Reconstructing the ancestral germline methylation state of young repeats. Mol Biol Evol 470:1777–1784
FitzGerald PC, Sturgill D, Shyakhtenko A, Oliver B, Vinson C (2006) Comparative genomics of Drosophila and human core promoters. Genome Biol 7:R53
Gertz EM, Yu YK, Agarwala R, Schäffer AA, Altschul SF (2006) Composition-based statistics and translated nucleotide searches: improving the TBLASTN module of BLAST. BMC Biol 4:41
Goll MG, Bestor TH (2005) Eukaryotic cytosine methyltransferases. Annu Rev Biochem 74:481–514
Gregory TR (2010) Animal genome size database. http://www.genomesize.com/
Grunau C, Renault E, Rosenthal A, Roizes G (2001) MethDB–a public database for DNA methylation data. Nucleic Acids Res 29:270–274. http://www.methdb.de
Guindon S, Gascuel O (2003) A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52:696–704
Hardeland U, Bentele M, Jiricny J, Schär P (2003) The versatile thymine DNA-glycosylase: a comparative characterization of the human, Drosophila and fission yeast orthologs. Nucleic Acids Res 31:2261–2271
Hejnol A, Obst M, Stamatakis A, Ott M, Rouse GW, Edgecombe GD, Martinez P, Baguñà J, Bailly X, Jondelius U, Wiens M, Müller WEG, Seaver E, Wheeler WC, Martindale MQ, Giribet G, Dunn CW (2009) Assessing the root of bilaterian animals with scalable phylogenomic methods. Proc Biol Sci 276:4261–4270
Ho KL, McNae IW, Schmiedeberg L, Klose RJ, Bird AP, Walkinshaw MD (2008) MeCP2 binding to DNA depends upon hydration at methyl-CpG. Mol Cell 29:525–531
Hodges C, Bintu L, Lubkowska L, Kashlev M, Bustamante C (2009) Nucleosomal fluctuations govern the transcription dynamics of RNA polymerase II. Science 325:626–628
Holterman M, van der Wurff A, van den Elsen S, van Megen H, Bongers T, Holovachov O, Bakker J, Helder J (2006) Phylum-wide analysis of SSU rDNA reveals deep phylogenetic relationships among nematodes and accelerated evolution toward crown clades. Mol Biol Evol 23:1792–1800
Hu TT, Pattyn P, Bakker EG, Cao J, Cheng JF, Clark RM, Fahlgren N, Fawcett JA, Grimwood J, Gundlach H, Haberer G, Hollister JD, Ossowski S, Ottilar RP, Salamov AA, Schneeberger K, Spannagl M, Wang X, Yang L, Nasrallah ME, Bergelson J, Carrington JC, Gaut BS, Schmutz J, Mayer KFX, Peer YVD, Grigoriev IV, Nordborg M, Weigel D, Guo YL (2011) The Arabidopsis lyrata genome sequence and the basis of rapid genome size change. Nat Genet 43:476–481
Jurka J, Kapitonov VV, Pavlicek A, Klonowski P, Kohany O, Walichiewicz J (2005) Repbase Update, a database of eukaryotic repetitive elements. Cytogenet Genome Res 110:462–467
Karlin S, Cardon LR (1994) Computational DNA sequence analysis. Annu Rev Microbiol 48:619–654
Kondrashov AS (2003) Direct estimates of human per nucleotide mutation rates at 20 loci causing Mendelian diseases. Hum Mutat 21:12–27
Krauss V, Eisenhardt C, Unger T (2009) The genome of the stick insect Medauroidea extradentata is strongly methylated within genes and repetitive DNA. PLoS ONE 4:e7223
Krauss V, Reuter G (2011) DNA methylation in Drosophila—a critical evaluation. Prog Mol Biol Transl Sci 101:177–191
Kriegs JO, Churakov G, Kiefmann M, Jordan U, Brosius J, Schmitz J (2006) Retroposed elements as archives for the evolutionary history of placental mammals. PLoS Biol 4:e91
Laurent L, Wong E, Li G, Huynh T, Tsirigos A, Ong CT, Low HM, Kin Sung KW, Rigoutsos I, Loring J, Wei CL (2010) Dynamic changes in the human methylome during differentiation. Genome Res 20:320–331
Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, Edsall L, Antosiewicz-Bourget J, Stewart R, Ruotti V, Millar AH, Thomson JA, Ren B, Ecker JR (2009) Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 462:315–322
Lynch M (2006) The origins of eukaryotic gene structure. Mol Biol Evol 23:450–468
Lynch M (ed) (2007) The origins of genome architecture. Sinauer Associates, Sunderland
Maddison WP, Maddison DR (2001) Mesquite: a modular system for evolutionary analysis. http://mesquiteproject.org
Maksakova IA, Romanish MT, Gagnier L, Dunn CA, van de Lagemaat LN, Mager DL (2006) Retroviral elements and their hosts: insertional mutagenesis in the mouse germ line. PLoS Genet 2:e2
Marhold J, Rothe N, Pauli A, Mund C, K K, Brueckner B, Lyko F (2004) Conservation of DNA methylation in dipteran insects. Insect Mol Biol 13:117–123
Maunakea AK, Nagarajan RP, Bilenky M, Ballinger TJ, D’Souza C, Fouse SD, Johnson BE, Hong C, Nielsen CB, Zhao Y, Turecki G, Delaney A, Varhol R, Thiessen N, Shchors K, Heine VM, Rowitch DH, Xing X, Fiore C, Schillebeeckx M, Jones SJM, Haussler D, Marra MA, Hirst M, Wang T, Costello JF (2010) Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature 466:253–257
Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB, Gnirke A, Jaenisch R, Lander ES (2008) Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature 454:766–770
Midford PE, Garland T, Maddison WP (2010) PDAP:PDTREE: a translation of the PDTREE application of Garland et al.’s phenotypic diversity analysis programs, version1.14
Molaro A, Hodges E, Fang F, Song Q, McCombie WR, Hannon GJ, Smith AD (2011) Sperm methylation profiles reveal features of epigenetic inheritance and evolution in primates. Cell 146:1029–1041
Morgan MT, Bennett MT, Drohat AC (2007) Excision of 5-halogenated uracils by human thymine DNA glycosylase. robust activity for DNA contexts other than cpg. J Biol Chem 282:27578–27586
Nanty L, Carbajosa G, Heap GA, Ratnieks F, van Heel DA, Down TA, Rakyan VK (2011) Comparative methylomics reveals gene-body H3K36me3 in Drosophila predicts DNA methylation and CpG landscapes in other invertebrates. Genome Res 21:1841–1850
Neafsey DE, Palumbi SR (2003) Genome size evolution in pufferfish: a comparative analysis of diodontid and tetraodontid pufferfish genomes. Genome Res 13:821–830
Okamura K, Matsumoto KA, Nakai K (2010) Gradual transition from mosaic to global DNA methylation patterns during deuterostome evolution. BMC Bioinforma 11(Suppl 7):S2
Popp C, Dean W, Feng S, Cokus SJ, Andrews S, Pellegrini M, Jacobsen SE, Reik W (2010) Genome-wide erasure of DNA methylation in mouse primordial germ cells is affected by AID deficiency. Nature 463:1101–1115
Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, Glöckner FO (2007) SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res 35:7188–7196
Rai K, Huggins IJ, James SR, Karpf AR, Jones DA, Cairns BR (2008) DNA demethylation in zebrafish involves the coupling of a deaminase, a glycosylase, and gadd45. Cell 135:1201–1212
Regev A, Lamb M, Jablonka E (1998) The role of DNA methylation in invertebrates: Developmental regulation or genome defense? Mol Biol Evol 15:880–891
Robinson KL, Tohidi-Esfahani D, Lo N, Simpson SJ, Sword GA (2011) Evidence for widespread genomic methylation in the migratory locust, Locusta migratoria (Orthoptera: Acrididae). PLos One 6:e28167
Rogozin IB, Iyer LM, Liang L, Glazko GV, Liston VG, Pavlov YI, Aravind L, Pancer Z (2007) Evolution and diversification of lamprey antigen receptors: evidence for involvement of an AID-APOBEC family cytosine deaminase. Nat Immunol 8:647–656
Rollins RA, Haghighi F, Edwards JR, Das R, Zhang MQ, Ju J, Bestor TH (2006) Large-scale structure of genomic methylation patterns. Genome Res 16:157–163
Sasai N, Defossez PA (2010) Many paths to one goal? The proteins that recognize methylated DNA in eukaryotes. Int J Dev Biol 53:323–334
Shoemaker R, Deng J, Wang W, Zhang K (2010) Allele-specific methylation is prevalent and is contributed by CpG-SNPs in the human genome. Genome Res 20:883–889
Simmen MW (2008) Genome-scale relationships between cytosine methylation and dinucleotide abundances in animals. Genomics 92:33–40
Simmen MW, Leitgeb S, Charlton J, Jones SJ, Harris BR, Clark VH, Bird A (1999) Nonmethylated transposable elements and methylated genes in a chordate genome. Science 283:1164–1167
Suzuki MM, Bird AP (2008) Dna methylation landscapes: provocative insights from epigenomics. Nat Rev Genet 9:465–476
Thalhammer A, Hansen AS, El-Sagheer AH, Brown T, Schofield CJ (2011) Hydroxylation of methylated CpG dinucleotides reverses stabilisation of DNA duplexes by cytosine 5-methylation. Chem Commun Camb 47:5325–5327
The Human Genome International Sequencing Consortium (2001) Initial sequencing and analysis of the human genome. Nature 409:860–921
Varriale A, Bernardi G (2006a) DNA methylation and body temperature in fishes. Gene 385:111–121
Varriale A, Bernardi G (2006b) DNA methylation in reptiles. Gene 385:122–127
Walsh CP, Bestor TH (1999) Cytosine methylation and mammalian development. Genes Dev 13:26–34
Walsh TK, Brisson JA, Robertson HM, Gordon K, Jaubert-Possamai S, Tagu D, Edwards OR (2010) A functional dna methylation system in the pea aphid, Acyrthosiphon pisum. Insect Mol Biol 19 Suppl 2:215–228
Wang Y, Jorda M, Jones PL, Maleszka R, Ling X, Robertson HM, Mizzen CA, Peinado MA, Robinson GE (2006) Functional CpG methylation system in a social insect. Science 314:645–647
Wu SC, Zhang Y (2010) Active DNA demethylation: many roads lead to rome. Nat Rev Mol Cell Biol 11:607–620
Yoder JA, Walsh CP, Bestor TH (1997) Cytosine methylation and the ecology of intragenomic parasites. Trends Genet 13:335–340
Zemach A, McDaniel IE, Silva P, Zilberman D (2010) Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science 328:916–919
Acknowledgments
This work was supported in part by DFG GRK-1384 and MA5082/1-1.
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Supplementary figures are available from the journal website. Machine readable supplementary data, including methylation rate data, genome size data, tree and the used genome versions are available at http://www.bioinf.uni-leipzig.de/supplements/10-026
Rights and permissions
About this article
Cite this article
Lechner, M., Marz, M., Ihling, C. et al. The correlation of genome size and DNA methylation rate in metazoans. Theory Biosci. 132, 47–60 (2013). https://doi.org/10.1007/s12064-012-0167-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12064-012-0167-y