Abstract
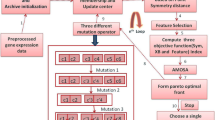
Studying the patterns hidden in gene expression data helps to understand the functionality of genes. But due to the large volume of genes and the complexity of biological networks it is difficult to study the resulting mass of data which often consists of millions of measurements. In order to reveal natural structures and to identify interesting patterns from the given gene expression data set, clustering techniques are applied. Semi-supervised classification is a new direction of machine learning. It requires huge unlabeled data and a few labeled data. Semi-supervised classification in general performs better than unsupervised classification. But to the best of our knowledge there are no works for solving gene expression data clustering problem using semi-supervised classification techniques. In the current paper we have made an attempt to solve the gene expression data clustering problem using a multiobjective optimization based semi-supervised classification technique with the aim to attain good quality partitions by using few labeled data. In order to generate the labeled data, initially Fuzzy C-means clustering technique is applied. In order to automatically determine the partitioning, multiple cluster centers corresponding to a cluster are encoded in the form of a string. In order to compute the quality of the obtained partitioning, values of five objective functions are computed. The effectiveness of this proposed semi-supervised clustering technique is demonstrated on five publicly available benchmark gene expression data sets. Comparison results with the existing techniques for gene expression data clustering prove that the proposed method is the most effective one. Statistical and biological significance tests have also been carried out.







Similar content being viewed by others
References
Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, Boldrick JC, Sabet H, Tran T, Yu X et al (2000) Distinct types of diffuse large b-cell lymphoma identified by gene expression profiling. Nature 403(6769):503–511
Alon U, Barkai N, Notterman DA, Gish K, Ybarra S, Mack D, Levine AJ (1999) Broad patterns of gene expression revealed by clustering analysis of tumor and normal colon tissues probed by oligonucleotide arrays. Proceed National Acad Sci 96(12):6745–6750
Altun Y, Belkin M, Mcallester DA (2005) Maximum margin semi-supervised learning for structured variables. In: Advances in neural information processing systems, pp 33–40
Bandyopadhyay S (2007) Analysis of biological data: a soft computing approach, World Scientific
Bandyopadhyay S, Saha S (2008) A point symmetry-based clustering technique for automatic evolution of clusters. Knowl Data Eng IEEE Trans 20(11):1441–1457
Bandyopadhyay S, Pal SK, Aruna B (2004) Multiobjective gas, quantitative indices, and pattern classification. Syst Man Cybern Part B Cybern IEEE Trans 34(5):2088–2099
Bandyopadhyay S, Mukhopadhyay A, Maulik U (2007) An improved algorithm for clustering gene expression data. Bioinformatics 23(21):2859–2865
Bandyopadhyay S, Saha S, Maulik U, Deb K (2008) A simulated annealing-based multiobjective optimization algorithm: AMOSA. Evol Comput IEEE Trans 12(3):269–283
Basu S, Banerjee A, Mooney RJ (2004) Active semi-supervision for pairwise constrained clustering
Bezdek JC (1981) Pattern recognition with fuzzy objective function algorithms. Kluwer Academic Publishers
Bilenko M, Basu S, Mooney RJ (2004) Integrating constraints and metric learning in semi-supervised clustering. In: Proceedings of the twenty-first international conference on Machine learning, ACM, pp 81–88
Brazma A, Vilo J (2000) Gene expression data analysis. FEBS lett 480(1):17–24
Brunet JP, Tamayo P, Golub TR, Mesirov JP (2004) Metagenes and molecular pattern discovery using matrix factorization. Proceed Natl Acad Sci 101(12):4164–4169
Chapelle O, Zien A (2004) Semi-supervised classification by low density separation. In AI STATS
Chapelle O, Schölkopf B, Zien A, et al. (2006) Semi-supervised learning, MIT press Cambridge
Cho RJ, Campbell MJ, Winzeler EA, Steinmetz L, Conway A, Wodicka L, Wolfsberg TG, Gabrielian AE, Landsman D, Lockhart DJ et al (1998) A genome-wide transcriptional analysis of the mitotic cell cycle. Mol cell 2(1):65–73
Chu S, DeRisi J, Eisen M, Mulholland J, Botstein D, Brown PO, Herskowitz I (1998) The transcriptional program of sporulation in budding yeast. Science 282(5389):699–705
De Smet F, Mathys J, Marchal K, Thijs G, De Moor B, Moreau Y (2002) Adaptive quality-based clustering of gene expression profiles. Bioinformatics 18(5):735–746
Deb K, Pratap A, Agarwal S, Meyarivan T, Fast A (2002) Nsga-ii. IEEE Trans Evol Comput 6(2):182–197
Dembélé D (2008) Multi-objective optimization for clustering 3-way gene expression data. Adv Data Anal Cl 2(3):211–225
Dhaeseleer P, Wen X, Fuhrman S, Somogyi R (1998) Mining the gene expression matrix: Inferring gene relationships from large scale gene expression data. In: Information processing in cells and tissues, Springer, pp 203–212
Eisen MB, Spellman PT, Brown PO, Botstein D (1998) Cluster analysis and display of genome-wide expression patterns. Proceed Natl Acad Sci 95(25):14863–14868
Everitt B (1974/1993) Cluster analysis. Halsted Press
Faceli K, de Souto MC, de Araújo DS, de Carvalho AC (2009) Multi-objective clustering ensemble for gene expression data analysis. Neurocomputing 72(13):2763–2774
Fraley C, Raftery AE (1998) How many clusters? which clustering method? answers via model-based cluster analysis. Comput J 41(8):578–588
Geman S, Geman D (1984) Stochastic relaxation, gibbs distributions, and the bayesian restoration of images. Patt Anal Mach Intell IEEE Trans 6:721–741
Ghosh D, Chinnaiyan AM (2002) Mixture modelling of gene expression data from microarray experiments. Bioinformatics 18(2):275–286
Herwig R, Poustka AJ, Müller C, Bull C, Lehrach H, O’Brien J (1999) Large-scale clustering of cdna-fingerprinting data. Genome Res 9(11):1093–1105
Heyer LJ, Kruglyak S, Yooseph S (1999) Exploring expression data: identification and analysis of coexpressed genes. Genome Res 9(11):1106–1115
Hu Q, Pan W, An S, Ma P, Wei J (2010) An efficient gene selection technique for cancer recognition based on neighborhood mutual information. Int J Mach Learn Cybern 1(1–4):63–74
Ishibuchi H, Murata T (1998) A multi-objective genetic local search algorithm and its application to flowshop scheduling. Syst Man Cybern Part C Appl Rev IEEE Trans 28(3):392–403
Iyer VR, Eisen MB, Ross DT, Schuler G, Moore T, Lee JC, Trent JM, Staudt LM, Hudson J, Boguski MS et al (1999) The transcriptional program in the response of human fibroblasts to serum. Science 283(5398):83–87
Jain AK, Murty MN, Flynn PJ (1999) Data clustering: a review. ACM Comput Surv 31(3):264–323
Jiang D, Pei J, Zhang A (2003) Dhc: a density-based hierarchical clustering method for time series gene expression data. In: Proceedings of Bioinformatics and Bioengineering. Third IEEE Symposium, pp 393–400
Jiang D, Tang C, Zhang A (2004) Cluster analysis for gene expression data: a survey. Knowl Data Eng IEEE Trans 16(11):1370–1386
Kirkpatrick S, Gelatt CD, Vecchi MP et al (1983) Optimization by simmulated annealing. Science 220(4598):671–680
Liu L, Hawkins DM, Ghosh S, Young SS (2003) Robust singular value decomposition analysis of microarray data. Proceed Natl Acad Sci 100(23):13,167–13,172
Lockhart D, Dong H, Byrne M, Follettie M, Gallo M, Chee M, Mittmann M, Wang C, Kobayashi M, Horton H et al (1996) Expression monitoring by hybridization to high-density oligonucleotide arrays. Nature Biotech 14(13):1675–1680
Lockhart DJ, Winzeler EA (2000) Genomics, gene expression and dna arrays. Nature 405(6788):827–836
Maulik U, Bandyopadhyay S (2002) Performance evaluation of some clustering algorithms and validity indices. Patt Anal Mach Intell IEEE Trans 24(12):1650–1654
Maulik U, Bandyopadhyay S (2003) Fuzzy partitioning using a real-coded variable-length genetic algorithm for pixel classification. Geosci Remote Sens IEEE Trans 41(5):1075–1081
Maulik U, Mukhopadhyay A, Bandyopadhyay S (2009) Combining pareto-optimal clusters using supervised learning for identifying co-expressed genes. BMC Bioinform 10(1):27
Mukhopadhyay A, Bandyopadhyay S, Maulik U (2010) Multi-class clustering of cancer subtypes through svm based ensemble of pareto-optimal solutions for gene marker identification. PloS one 5(11):e13803
Mukhopadhyay A, Maulik U, Bandyopadhyay S (2013) An interactive approach to multiobjective clustering of gene expression patterns. Biomed Eng IEEE Trans 60(1):35–41
Qin ZS (2006) Clustering microarray gene expression data using weighted chinese restaurant process. Bioinformatics 22(16):1988–1997
Reymond P, Weber H, Damond M, Farmer EE (2000) Differential gene expression in response to mechanical wounding and insect feeding in arabidopsis. Plant Cell Online 12(5):707–719
Rose K (1998) Deterministic annealing for clustering, compression, classification, regression, and related optimization problems. In: Proceedings of the IEEE 86(11):2210–2239
Rousseeuw PJ (1987) Silhouettes: a graphical aid to the interpretation and validation of cluster analysis. J Comput Appl Math 20:53–65
Saha S, Bandyopadhyay S (2013) A generalized automatic clustering algorithm in a multiobjective framework. Appl Soft Comput 13(1):89–108
Saha S, Ekbal A, Alok AK (2012) Semi-supervised clustering using multiobjective optimization. In: Hybrid Intelligent Systems (HIS), 12th International Conference, IEEE, pp 360–365
Saha S, Ekbal A, Gupta K, Bandyopadhyay S (2013) Gene expression data clustering using a multiobjective symmetry based clustering technique. Comput Biol Med 43(11):1965–1977
Santos JM, Embrechts M (2009) On the use of the adjusted rand index as a metric for evaluating supervised classification. In: Artificial Neural Networks-ICANN, Springer, pp 175–184
Schena M, Shalon D, Davis RW, Brown PO (1995) Quantitative monitoring of gene expression patterns with a complementary dna microarray. Science 270(5235):467–470
Schott JR (1995) Fault tolerant design using single and multicriteria genetic algorithm optimization. Tech Rep DTIC Doc
Sharan R, Shamir R (2000) Click: a clustering algorithm with applications to gene expression analysis. Proceed Int Conf Intell Syst Mol Biol 8:16
Sharma A, Imoto S, Miyano S, Sharma V (2012) Null space based feature selection method for gene expression data. Int J Mach Learn Cybern 3(4):269–276
Sherlock G (2000) Analysis of large-scale gene expression data. Curr Opin Immunol 12(2):201–205
de Souto MC, Costa IG, de Araujo DS, Ludermir TB, Schliep A (2008) Clustering cancer gene expression data: a comparative study. BMC Bioinform 9(1):497
Tamayo P, Slonim D, Mesirov J, Zhu Q, Kitareewan S, Dmitrovsky E, Lander ES, Golub TR (1999) Interpreting patterns of gene expression with self-organizing maps: methods and application to hematopoietic differentiation. Proceed Natl Acad Sci 96(6):2907–2912
Tang VT, Yan H (2012) Noise reduction in microarray gene expression data based on spectral analysis. Int J Mach Learn Cybern 3(1):51–57
Tavazoie S, Hughes JD, Campbell MJ, Cho RJ, Church GM (1999) Systematic determination of genetic network architecture. Nature Genet 22(3):281–285
Tou JT GR (1974) Pattern recognition principles. Reading: Addison-Wesley
Troyanskaya O, Cantor M, Sherlock G, Brown P, Hastie T, Tibshirani R, Botstein D, Altman RB (2001) Missing value estimation methods for dna microarrays. Bioinformatics 17(6):520–525
Wen X, Fuhrman S, Michaels GS, Carr DB, Smith S, Barker JL, Somogyi R (1998) Large-scale temporal gene expression mapping of central nervous system development. Proceed Natl Acad Sci 95(1):334–339
Wilcoxon F, Katti S, Wilcox RA (1963) Critical values and probability levels for the Wilcoxon rank sum test and the Wilcoxon signed rank test. American Cyanamid Comp
Xie XL, Beni G (1991) A validity measure for fuzzy clustering. IEEE Trans Patt Anal Mach Intell 13(8):841–847
Xu X (2013) Enhancing gene expression clustering analysis using tangent transformation. Int J Mach Learn Cybern 4(1):31–40
Yeung KY, Fraley C, Murua A, Raftery AE, Ruzzo WL (2001) Model-based clustering and data transformations for gene expression data. Bioinformatics 17(10):977–987
Zitzler E, Laumanns M, Thiele L (2001) Spea 2: Improving the strength pareto evolutionary algorithm
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Alok, A.K., Saha, S. & Ekbal, A. Semi-supervised clustering for gene-expression data in multiobjective optimization framework. Int. J. Mach. Learn. & Cyber. 8, 421–439 (2017). https://doi.org/10.1007/s13042-015-0335-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13042-015-0335-8