Abstract
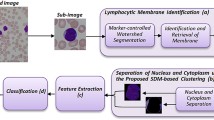
The goal of Big Data analysis is delineating hidden patterns from data and leverage them into strategies and plans to support informed decision making in a diversity of situations. Big Data are characterized by large volume, high velocity, wide variety, and high value, which may represent difficulties in storage and processing. Research on Big Data repositories has contributed promising results that primarily address how to efficiently mine a variety of large volume of structured and unstructured data. However, innovative insights can emerge while leveraging the value characteristic of Big Data. In other words, any given data can be big if analytics can draw a big value from it. In this paper, we demonstrate the potential of five machine learning algorithms to leverage the value of medium size microscopic blood smear images to classify patients with chronic lymphocytic leukemia (CLL). The maximum majority voting method is used to fuse the predications made by the five classifier models. To validate this work, 11 CLL patients are refereed by flow cytometry equipment and the results are compared to the proposed classifier model. The proposed method proceeds through a sequence of steps while working with the lymphocyte images: it segments the lymphocyte images, extracts/selects features, classifies the selected features using five classifiers, and calculates the majority class for the test image. The proposed composite classifier model has an accuracy of 87.0%, true-positive rate of 84.95%, and 10.96% false-positive rate and can correctly identify 9 out of 11 patients as positive for CLL.









Similar content being viewed by others
References
Abdul Nasir A, Mashor M, Hassan R (2012) Leukaemia screening based on fuzzy ARTMAP and simplified fuzzy ARTMAP neural networks. In: 2012 IEEE EMBS conference on biomedical engineering and sciences (IECBES), IEEE, pp 11–16
Adjouadi M, Zong N, Ayala M (2005) Multidimensional pattern recognition and classification of white blood cells using support vector machines. Part Part Syst Charact 22:107–118
Allab K, Labiod L, Nadif M (2017) A semi-NMF-PCA unified framework for data clustering. IEEE Trans Knowl Data Eng 29:2–16
Alpaydin E (2007) Combining pattern classifiers: methods and algorithms (kuncheva, li; 2004) [book review]. IEEE Trans Neural Netw 18:964
Bain BJ (2008) A beginner’s guide to blood cells, 2nd edn. Wiley, San Francisco
Burbidge R, Rowland JJ, King RD (2007) Active learning for regression based on query by committee. In: International conference on intelligent data engineering and automated learning. Springer, pp 209–218
Calgary Laboratory Services (2016) https://www.calgarylabservices.com/. Accessed 30 Dec 2016
Canadian Cancer Society (2016) http://www.cancer.ca/. Accessed 30 Dec 2016
Canadian Cancer Statistics (2016) http://www.cancer.ca/~/media/cancer.ca/CW/cancer%20information/cancer%20101/Canadian%20cancer%20statistics/canadian-cancer-statistics-2013-EN.pdf. Accessed 30 Dec 2016
CellaVision Company (2016) http://www.cellavision.com. Accessed 08 Dec 2016
Chen T-T (2016) Predicting analysis times in randomized clinical trials with cancer immunotherapy. BMC Med Res Methodol 16:1
Chen W-P, Hung C-L, Tsai S-JJ, Lin Y-L (2014) Novel and efficient tag SNPs selection algorithms. Bio-Med Mater Eng 24:1383–1389
Clinton N, Holt A, Yan L, Gong P (2008) An accuracy assessment measure for object based image segmentation. Int Arch Photogramm Remote Sens Spat Inf Sci 37:1189–1194
Craig FE, Foon KA (2008) Flow cytometric immunophenotyping for hematologic neoplasms. Blood 111:3941–3967
Dai L, Gao X, Guo Y, Xiao J, Zhang Z (2012) Bioinformatics clouds for big data manipulation. Biol Direct 7:43
Feature Selection Software Component (2016) http://www.mathworks.com/matlabcentral/fileexchange/22970-feature-selection-using-matlab. Accessed 21 Dec 2016
Freeman C, Kulić D, Basir O (2015) An evaluation of classifier-specific filter measure performance for feature selection. Pattern Recognit 48:1812–1826
Freund Y, Schapire RE (1995) A desicion-theoretic generalization of on-line learning and an application to boosting. In: European conference on computational learning theory. Springer, pp 23–37
Fu Y, Zhu X, Elmagarmid AK (2013) Active learning with optimal instance subset selection. IEEE Trans Cybern 43:464–475
Fukunaga K (1990) Introduction to statistical pattern recognition, 1st edn. Academic, San Diego
Gould N, Toint PL (2004) Preprocessing for quadratic programming. Math Program 100:95–132
Grever MR et al (2007) Comprehensive assessment of genetic and molecular features predicting outcome in patients with chronic lymphocytic leukemia: results from the US Intergroup Phase III Trial E2997. J Clin Oncol 25:799–804
Guo N, Zeng L, Wu Q (2007) A method based on multispectral imaging technique for white blood cell segmentation. Comput Biol Med 37:70–76
Healey R, Patel JL, de Koning L, Naugler C (2015) Incidence of chronic lymphocytic leukemia and monoclonal B-cell lymphocytosis in Calgary, Alberta, Canada. Leuk Res 39:429–434
Herring W, Pearson I, Purser M, Nakhaipour HR, Haiderali A, Wolowacz S, Jayasundara K (2016) Cost effectiveness of ofatumumab plus chlorambucil in first-line chronic lymphocytic leukaemia in Canada. PharmacoEconomics 34:77–90
Houwen B (2001) The differential cell count. Lab Hematol 7:89–100
Hsu C-W, Chang C-C, Lin C-J (2003) A practical guide to support vector classification. Data Sci Assoc 1–16
Hu Z, Bao Y, Xiong T, Chiong R (2015) Hybrid filter–wrapper feature selection for short-term load forecasting. Eng Appl Artif Intell 40:17–27
Jaffar MA, Ishtiaq M, Ahmed B (2010) Fuzzy wavelet-based color image segmentation using self-organizing neural network. Intern J Innov Comput Inf Control (IJICIC) 6(11):4813–4824
Jiang K, Liao Q-M, Xiong Y (2006) A novel white blood cell segmentation scheme based on feature space clustering. Soft Comput 10:12–19
Kaplan RS, Porter ME (2011) How to solve the cost crisis in health care. Harv Bus Rev 89:46–52
Ko BC, Gim J-W, Nam J-Y (2011) Automatic white blood cell segmentation using stepwise merging rules and gradient vector flow snake. Micron 42:695–705
Kohlwey E, Sussman A, Trost J, Maurer A (2011) Leveraging the cloud for big data biometrics: meeting the performance requirements of the next generation biometric systems. In: 2011 IEEE World Congress on Services (SERVICES), IEEE, pp 597–601
Lagarias JC, Reeds JA, Wright MH, Wright PE (1998) Convergence properties of the Nelder–Mead simplex method in low dimensions. SIAM J Optim 9:112–147
Lawson CL, Hanson RJ (1974) Solving least squares problems, vol 161. SIAM, Philadelphia, PA, USA
Loh WY (2011) Classification and regression trees. Wiley Interdiscip Rev Data Min Knowl Discov 1:14–23
Lorena AC, de Carvalho AC (2005) Minimum spanning trees in hierarchical multiclass support vector machines generation. In: Ali M, Esposito F (eds) Innovations in applied artificial intelligence. Springer, pp 422–431
Madhloom H, Kareem S, Ariffin H, Zaidan A, Alanazi H, Zaidan B (2010) An automated white blood cell nucleus localization and segmentation using image arithmetic and automatic threshold. J Appl Sci 10:959–966
Madhloom HT, Kareem SA, Ariffin H (2012) An image processing application for the localization and segmentation of lymphoblast cell using peripheral blood images. J Med Syst 36:2149–2158
Mathews JD et al (2013) Cancer risk in 680 000 people exposed to computed tomography scans in childhood or adolescence: data linkage study of 11 million Australians. BMJ: Br Med J 346(10):1–18
McPherson RA, Pincus MR (2011) Henry’s clinical diagnosis and management by laboratory methods, 22nd edn. Elsevier Health Sciences, Philadelphia
Mohammed E, Mohamed M, Naugler C, Far B (2013) Application of support vector machine and k-means clustering algorithms for robust chronic lymphocytic leukemia color cell segmentation. In: Proceedings of the 15th IEEE international conference on e-Health Networking, Application and Services HEALTHCOM, Lisbon. IEEE, pp 622–626. doi:10.1109/HealthCom.2013.6720751
Musen MA, Middleton B, Greenes RA (2014) Clinical decision-support systems. In: Shortliffe EH, Cimino JJ (eds) Biomedical informatics. Springer, pp 643–674
Oliai C (2013) Small lymphocytic lymphoma. In: Brady LW, Yaeger TE (eds) Encyclopedia of radiation oncology. Springer, pp 798–798
Otsu N (1975) A threshold selection method from gray-level histograms. Automatica 11:23–27
Rajaraman A, Ullman JD (2012) Mining of massive datasets. Cambridge University Press, Cambridge, United Kingdom
Ramoser H (2008) Leukocyte segmentation and SVM classification in blood smear images. Mach Graph Vis Int J 17:187–200
Reta C, Robles LA, Gonzalez JA, Diaz R, Guichard JS (2010) Segmentation of bone marrow cell images for morphological classification of acute leukemia. In: FLAIRS Conference
Ripley B (2002) Statistical data mining. Springer, New York
Rothwell PM et al (2012) Short-term effects of daily aspirin on cancer incidence, mortality, and non-vascular death: analysis of the time course of risks and benefits in 51 randomised controlled trials. Lancet 379:1602–1612
Sabino DMU, Costa LDF, Rizzatti E, Zago M (2004) Toward leukocyte recognition using morphometry, texture and color. In: IEEE international symposium on biomedical imaging: nano to macro. IEEE, pp 121–124
Sadeghian F, Seman Z, Ramli AR, Kahar BA, Saripan M-I (2009) A framework for white blood cell segmentation in microscopic blood images using digital image processing. Biol Proced Online 11:196–206
Seftel M et al (2009) High incidence of chronic lymphocytic leukemia (CLL) diagnosed by immunophenotyping: a population-based Canadian cohort. Leuk Res 33:1463–1468
Shivhare S, Shrivastava R (2012) Morphological granulometric feature of nucleus in automatic bone marrow white blood cell classification. Int J Sci Res Publ 2:1–7
Sobajic O, Moussavi M, Far B (2010) Parameterized strategy pattern. In: Proceedings of the 17th conference on pattern languages of programs. ACM, p 9
Tam CS et al (2008) Chronic lymphocytic leukaemia CD20 expression is dependent on the genetic subtype: a study of quantitative flow cytometry and fluorescent in situ hybridization in 510 patients. Br J Haematol 141:36–40
The Language of Technical Computing (2016) http://www.mathworks.com/products/matlab/. Accessed 20 Dec 2016
Trigeorgis G, Bousmalis K, Zafeiriou S, Schuller B (2014) A deep semi-NMF model for learning hidden representations. In: ICML, pp 1692–1700
Ushizima DM, Lorena AC, De Carvalho A (2005) Support vector machines applied to white blood cell recognition. In: Fifth international conference on hybrid intelligent systems, 2005. HIS’05. IEEE, pp 6–11
Ververidis D, Kotropoulos C (2008) Fast and accurate sequential floating forward feature selection with the Bayes classifier applied to speech emotion recognition. Signal Process 88:2956–2970
Vollset SE et al (2013) Effects of folic acid supplementation on overall and site-specific cancer incidence during the randomised trials: meta-analyses of data on 50 000 individuals. Lancet 381:1029–1036
Wang K (2014) BioPig a Hadoop-based analytic toolkit for large scale sequence data. Bioinformatics 29(23):3014–3019
Wang W, Haerian K, Salmasian H, Harpaz R, Chase H, Friedman C (2011) A drug-adverse event extraction algorithm to support pharmacovigilance knowledge mining from PubMed citations. In: AMIA annual symposium proceedings, 2011. American Medical Informatics Association, p 1464
Wang L, Chen D, Ranjan R, Khan SU, KolOdziej J, Wang J (2012a) Parallel processing of massive EEG data with MapReduce. In: ICPADS, pp 164–171
Wang X-Y, Zhang X-J, Yang H-Y, Bu J (2012b) A pixel-based color image segmentation using support vector machine and fuzzy C-means. Neural Netw 33:148–159
Wang Y, Wang J, Liao H, Chen H (2017) An efficient semi-supervised representatives feature selection algorithm based on information theory. Pattern Recognit 61:511–523
Xu X, Tsang IW, Xu D (2013) Soft margin multiple kernel learning. IEEE Trans Neural Netw Learn Syst 24:749–761
Yegnanarayana B (2006) Artificial neural networks, 1st edn. PHI Learning Pvt. Ltd., India Institute of Technology, New Delhi, India
Zhang Z, Bai L, Liang Y, Hancock E (2017a) Joint hypergraph learning and sparse regression for feature selection. Pattern Recognit 63:291–309
Zhang Z, Zhang Y, Li F, Zhao M, Zhang L, Yan S (2017b) Discriminative sparse flexible manifold embedding with novel graph for robust visual representation and label propagation. Pattern Recognit 61:492–510
Zhuang H, Low K-S, Yau W-Y (2012) Multichannel pulse-coupled-neural-network-based color image segmentation for object detection. IEEE Trans Ind Electron 59:3299–3308
Acknowledgements
This work has been supported and funded by SmartLabs Ltd., Calgary, AB, Canada and MITACS Accelerate program under Grant IT01892/FR02553.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Mohammed, E.A., Mohamed, M.M.A., Naugler, C. et al. Toward leveraging big value from data: chronic lymphocytic leukemia cell classification. Netw Model Anal Health Inform Bioinforma 6, 6 (2017). https://doi.org/10.1007/s13721-017-0146-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13721-017-0146-9