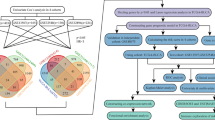
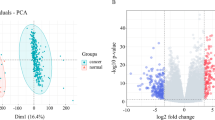
Abstract
Predicting bladder cancer outcomes is important for patients’ treatments, and it’s common to predict the outcomes from omics data. However, using a single type of omics data suffers from data noise since individual omics type represents only one single view of bladder cancer patients. In this study, we have estimated bladder cancer prognosis by integrating multi-omics data including RNA-seq, miRNA-seq, DNA methylation, and copy number variation data. To effectively integrate the multi-omics data, we have developed a transfer-learning based Cox proportional hazards network (TCAP) by utilizing an integrated loss function consisted of two modules: the data reconstruction module to ensure learning a representative hidden layer for the input data, and the proportional hazard module to estimate patients’ risks. The experiments on 336 patients from The Cancer Genome Atlas (TCGA) showed that our method achieved a concordance index (C-index) of 0.665, which is higher than previous methods. In consideration of the expense to obtain multi-omics data in clinics, we fitted the risks estimated from TCAP by training an XGboost model based on mRNA data only. The model achieved a reasonable C-index of 0.621, and independent tests on three additional datasets achieved an average C-index of 0.637 \(\pm\) 0.047. The essentially same result as the one achieved on TCGA dataset indicates the robustness of our model. Based on the risk subgroups divided by TCAP, we identified 12 candidate genes that affected the survival of bladder cancer patients, among which 7 genes (58.3%) have been proved to associate with bladder cancer through literature review. In summary, the results indicated that we have constructed an accurate and robust model for predicting bladder cancer outcomes.



Similar content being viewed by others
Availability of data and material
All the data analyzed during the current study are available in the TCGA dataset (https://tcga-data.nci.nih.gov/tcga/) and the GEO database (https://www.ncbi.nlm.nih.gov).
Code availability
The method codes are available at: https://github.com/Hua0113/TCAP.
References
Bokde, N., et al.: A novel imputation methodology for time series based on pattern sequence forecasting. Pattern Recognit. Lett. 116, 88–96 (2018)
Chai, H., et al.: Integrating multi-omics data through deep learning for accurate cancer prognosis prediction. Comput. Biol. Med. 2021, 104481 (2021)
Cheerla, A., Gevaert, O.: Deep learning with multimodal representation for pancancer prognosis prediction. Bioinformatics 35(14), i446–i454 (2019)
Chen, T., Guestrin, C.: Xgboost: a scalable tree boosting system. In: Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining. ACM; 2016, pp. 785–794 (2016)
Knowles, L.M., et al.: CLT1 targets bladder cancer through integrin α5β1 and CLIC3. Mol. Cancer Res. 11(2), 194–203 (2013)
Lee, T.-Y., et al.: Incorporating deep learning and multi-omics autoencoding for analysis of lung adenocarcinoma prognostication. Comput. Biol. 87, 107277 (2020)
Lin, J., et al.: A robust 11-genes prognostic model can predict overall survival in bladder cancer patients based on five cohorts. Cancer Cell Int. 20(1), 1–14 (2020)
Liu, A., et al.: Prognostic value of the combined expression of tumor-associated trypsin inhibitor (TATI) and p53 in patients with bladder cancer undergoing radical cystectomy. Cancer Biomark. 26(3), 281–289 (2019)
Lv, S., et al.: PPARγ activation serves as therapeutic strategy against bladder cancer via inhibiting PI3K–Akt signaling pathway. BMC Cancer 19(1), 1–13 (2019)
Mariette, J., Villa-Vialaneix, N.: Unsupervised multiple kernel learning for heterogeneous data integration. Bioinformatics 34(6), 1009–1015 (2018)
Noguchi, S., et al.: Replacement treatment with microRNA-143 and-145 induces synergistic inhibition of the growth of human bladder cancer cells by regulating PI3K/Akt and MAPK signaling pathways. Cancer Lett. 328(2), 353–361 (2013)
Ou, Y., et al.: Activation of cyclic AMP/PKA pathway inhibits bladder cancer cell invasion by targeting MAP4-dependent microtubule dynamics. Urol. Oncol. Semin. Orig. Investig. 32(1), 47–e21 (2014)
Richters, A., Aben, K.K., Kiemeney, L.A.: The global burden of urinary bladder cancer: an update. World J. Urol. 38(8), 1895–1904 (2020)
Ritchie, M.E., et al.: limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43(7), e47 (2015)
Rohart, F., et al.: mixOmics: an R package for ’omics feature selection and multiple data integration. PLoS Comput. Biol. 13(11), e1005752 (2017)
Salimian, J., et al.: MiR-486-5p enhances cisplatin sensitivity of human muscle-invasive bladder cancer cells by induction of apoptosis and down-regulation of metastatic genes. Urol. Oncol. Semin. Orig. Investig. 38(9), 738–e9 (2020)
Song, B.N., Chu, I.S.: A gene expression signature of FOXM1 predicts the prognosis of hepatocellular carcinoma. Exp. Mol. Med. 50(1), e418 (2018)
Stevens, T.A., et al.: Identification of novel binding elements and gene targets for the homeodomain protein BARX2. J. Biol. Chem. 279(15), 14520–14530 (2004)
Stirzaker, C., et al.: Methylome sequencing in triple-negative breast cancer reveals distinct methylation clusters with prognostic value. Nat. Commun. 6, 5899 (2015)
Tomczak, K., Czerwinska, P., Wiznerowicz, M.: The Cancer Genome Atlas (TCGA): an immeasurable source of knowledge. Contemp. Oncol. (pozn) 19(1A), A68-77 (2015)
Tong, D., et al.: Improving prediction performance of colon cancer prognosis based on the integration of clinical and multi-omics data. BMC Med. Inform. 20(1), 22 (2020a)
Tong, L., et al.: Deep learning based feature-level integration of multi-omics data for breast cancer patients survival analysis. BMC Med. Inform. 20(1), 1–12 (2020b)
Van Belle, V., et al.: Support vector methods for survival analysis: a comparison between ranking and regression approaches. Artif. Intell. Med. 53(2), 107–118 (2011)
Vanacker, H., et al.: Enhanced performance of prognostic estimation from TCGA RNAseq data using transfer learning. Ann. Oncol. 30, v52 (2019)
Volinia, S., Croce, C.M.: Prognostic microRNA/mRNA signature from the integrated analysis of patients with invasive breast cancer. Proc. Natl. Acad. Sci. USA 110(18), 7413–7417 (2013)
Volkmer, J.P., et al.: Three differentiation states risk-stratify bladder cancer into distinct subtypes. Proc. Natl. Acad. Sci. USA 109(6), 2078–2083 (2012)
Wei, L., et al.: TCGA-assembler 2: software pipeline for retrieval and processing of TCGA/CPTAC data. Bioinformatics 34(9), 1615–1617 (2018)
Xie, C., et al.: KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 39(3), W316-322 (2011)
Zhang, Y., et al.: Identification of core genes and key pathways via integrated analysis of gene expression and DNA methylation profiles in bladder cancer. Med. Sci. Monit. Int. Med. J. Exp. 24, 3024 (2018)
Funding
The work was supported in part by the National Key R&D Program of China (2020YFB0204803), National Natural Science Foundation of China (61772566 and 81801132), Guangdong Frontier & Key Tech Innovation Program (2018B010109006, 2019B020228001), Natural Science Foundation of Guangdong, China (2019A1515012207), and Introducing Innovative and Entrepreneurial Teams (2016ZT06D211).
Author information
Authors and Affiliations
Contributions
CH and YY conceived the study. CH and ZZ performed the data analysis. CH and WY interpreted the results. CH and ZZ wrote the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Ethics approval
Not applicable.
Consent to participate
All the authors listed have approved the manuscript.
Consent for publication
All the authors listed have approved the manuscript.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Chai, H., Zhang, Z., Wang, Y. et al. Predicting bladder cancer prognosis by integrating multi-omics data through a transfer learning-based Cox proportional hazards network. CCF Trans. HPC 3, 311–319 (2021). https://doi.org/10.1007/s42514-021-00074-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42514-021-00074-9