Abstract
Finding good feedback documents for query expansion is a well-known problem in the field of information retrieval. This paper describes a novel approach for finding relevant documents for feedback in query expansion for biomedical document retrieval. The proposed approach relies on a small amount of human intervention to find good feedback documents and tries to learn the relation between query and documents in terms of usefullness of document for query expansion. This proposed approach uses an NLP-based feature weighting technique with classification and clustering method on the documents and identifies relevant documents for feedback. The documents are represented using term frequency and inverse document frequency (TF–IDF) features and these features are weighted according to the type of query and type of the terms. The experiments performed on CDS 2014, 2015 and 2016 datasets show that the feature weighting in finding feedback documents for query expansion approach gives good results as compared to the results of pseudo-relevance feedback, relevance feedback and the results of TF–IDF features without weighting.


Similar content being viewed by others
Explore related subjects
Discover the latest articles and news from researchers in related subjects, suggested using machine learning.Notes
References
Allan J. Incremental relevance feedback for information filtering. In: Proceedings of the 19th annual international ACM SIGIR conference on research and development in information retrieval. pp. 270–278. ACM; 1996.
Aronson AR, Rindflesch TC. Query expansion using the UMLS metathesaurus. In: Proceedings of the AMIA annual fall symposium. p. 485. American Medical Informatics Association; 1997.
Azad HK, Deepak A. Query expansion techniques for information retrieval: a survey. Inf Process Manag. 2019;56(5):1698–735.
Boag W, Wacome K, Naumann T, Rumshisky A. Cliner: a lightweight tool for clinical named entity recognition. In: AMIA joint summits on clinical research informatics (poster); 2015.
Cao G, Nie JY, Gao J, Robertson S. Selecting good expansion terms for pseudo-relevance feedback. In: Proceedings of the 31st annual international ACM SIGIR conference on research and development in information retrieval. pp. 243–250. ACM; 2008.
Carpineto C, Romano G. A survey of automatic query expansion in information retrieval. ACM Comput Surv (CSUR). 2012;44(1):1.
Díaz-Galiano MC, Martín-Valdivia MT, Ureña-López L. Query expansion with a medical ontology to improve a multimodal information retrieval system. Comput Biol Med. 2009;39(4):396–403.
Dong L, Srimani PK, Wang JZ. Ontology graph based query expansion for biomedical information retrieval. In: Bioinformatics and biomedicine (BIBM), 2011 IEEE international conference on. pp. 488–493. IEEE; 2011.
Oh HS, Jung Y. Cluster-based query expansion using external collections in medical information retrieval. J Biomed Inform. 2015;58:70–9.
Pan M, Zhang Y, He T, Jiang X. An enhanced hal-based pseudo relevance feedback model in clinical decision support retrieval. In: International conference on intelligent computing. pp. 93–99. Springer; 2018.
Ramshaw LA, Marcus MP. Text chunking using transformation-based learning. Natural language processing using very large corpora. New York: Springer; 1999. p. 157–76.
Roberts K, Simpson M, Demner-Fushman D, Voorhees E, Hersh W. State-of-the-art in biomedical literature retrieval for clinical cases: a survey of the TREC 2014 CDS track. Inf Retr J. 2016;19(1–2):113–48.
Salton G, Buckley C. Improving retrieval performance by relevance feedback. J Am Soc Inf Sci. 1990;41(4):288–97.
Sankhavara J. Biomedical document retrieval for clinical decision support system. In: Proceedings of ACL 2018, student research workshop. pp. 84–90; 2018.
Sankhavara J, Majumder P. Biomedical information retrieval. In: Working notes of FIRE 2017—Forum for information retrieval evaluation. pp. 154–157; 2017.
Sankhavara J, Thakrar F, Sarkar S, Majumder P. Fusing manual and machine feedback in biomedical domain. Tech. rep., Dhirubhai Ambani Inst of Information and Communication Technology. 2014.
Stokes N, Li Y, Cavedon L, Zobel J. Exploring criteria for successful query expansion in the genomic domain. Inf Retr. 2009;12(1):17–50.
Uzuner Ö, South BR, Shen S, DuVall SL. 2010 i2b2/VA challenge on concepts, assertions, and relations in clinical text. J Am Med Inform Assoc. 2011;18(5):552–6.
Xu J, Croft WB. Quary expansion using local and global document analysis. In: ACM sigir forum. vol. 51, pp. 168–175. ACM; 2017.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Author has received research grant from TCS research scholar program.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the topical collection “Forum for Information Retrieval Evaluation” guest edited by Mandar Mitra and Prasenjit Majumder.
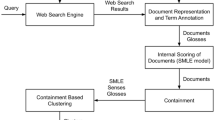
Feedback Document Discovery: Finding Good Feedback Documents
Feedback Document Discovery: Finding Good Feedback Documents
First Algorithm
The first algorithm is based on classification. If we have human judgements available for some of the feedback documents, then it will serve as a training data for classification. The documents are represented as a collection of bag-of-words, the TF–IDF scores of the words represent features and human relevance scores provides the classes. Using this as a training data, we want to predict the relevance of other top retrieved feedback documents represented by TF–IDF scores of words.

Second Algorithm
The second algorithm is an extension of first algorithm. The analysis of results of first algorithm shows that the feedback document set still contains some non-relevant docs and it is responsible for insignificant improvement. The detailed analysis is given in “Experiments and Results”. This approach further removes non-relevant documents from relevant document class identified by classification approach. The idea is to perform clustering on the relevant identified documents with number of clusters as two: one from actually relevant documents and second from non-relevant documents. This approach relies on the statement that the relevant documents tend to cluster within the space. k-means clustering is used with k = 2. Since, the convergence of k-means clustering depends on the initial choice of cluster centroids, the initial cluster centroids are chosen as the average of relevant documents’ vectors and the average of non-relevant documents’ vectors from training data.

Rights and permissions
About this article
Cite this article
Sankhavara, J. Feature Weighting in Finding Feedback Documents for Query Expansion in Biomedical Document Retrieval. SN COMPUT. SCI. 1, 75 (2020). https://doi.org/10.1007/s42979-020-0069-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s42979-020-0069-x